Effect of Vector Density and Competence on Macromolecular Vector Transformation Efficiency
,
1.Suzhou Industrial Park Institute of Services Outsourcing, Suzhou 215123, China; 2. Genewiz (Suzhou) Biotechnology Co., Ltd., Suzhou 215123, China
1 Introduction
The plasmid vector is the small DNA molecule having autonomous replication ability in cells, and the most common recombinant DNA carrier in genetic engineering. It can use the recombinant DNA technology to transport the purposefully strong DNA fragments into recipient cells for propagation and expression[1-7]. The DNA length of plasmid vector varies from a few thousand base pairs to hundreds of thousands of base pairs. Compared with the small molecular weight vector, the macromolecular vector has low extracellular recombination and connection efficiency, low transformation efficiency in competence, and low replication rate in the host cells, so it is easy to produce random mutation and deletion of target DNA and unstable expression in the passage[8-10]. Therefore, it needs an optimized recombination and connection system and an optimized competence transformation system, so as to improve the recombination and transformation efficiency of macromolecular vector[11-13]. In this study, the PCR (Polymerase Chain Reaction) technology was used to design a 835bp target gene synthesized from 26 primers, and this gene was connected with about 20 kb macromolecular vector pCMV5-Cla1-RyR1_WT to form the recombinant plasmid. On the basis of 1500 ng target gene substrate, the recombinant reaction of 6-gradient vector amount (50 ng, 100 ng, 150 ng, 200 ng, 250 ng and 300 ng) was carried out, and the recombinant reaction products obtained were transformed into 6 different kinds of competence (Top10F’, DH5α, Stbl3, Epi400, JM108, SCSI) to form 36 different combinations. Then the rate of positive clones from the 36 different combinations was analyzed, respectively, in order to get the optimal vector amount and best competence in the recombinant cloning of macromolecular vector.
2 Materials and methods
2.1Materials
2.1.1Target gene. In this study, we selected one 835 bp target gene (Fig. 1). The overall GC content was 56.17%, it was not the gene with high GC content, and there was slight fluctuation in the GC content. Overall, it was easy to be amplified (Fig. 2). The target gene had no significant direct, inverted repeat, or auto-reduplication, without difficulty in primer design.

Fig.1Completesequenceoftargetgene
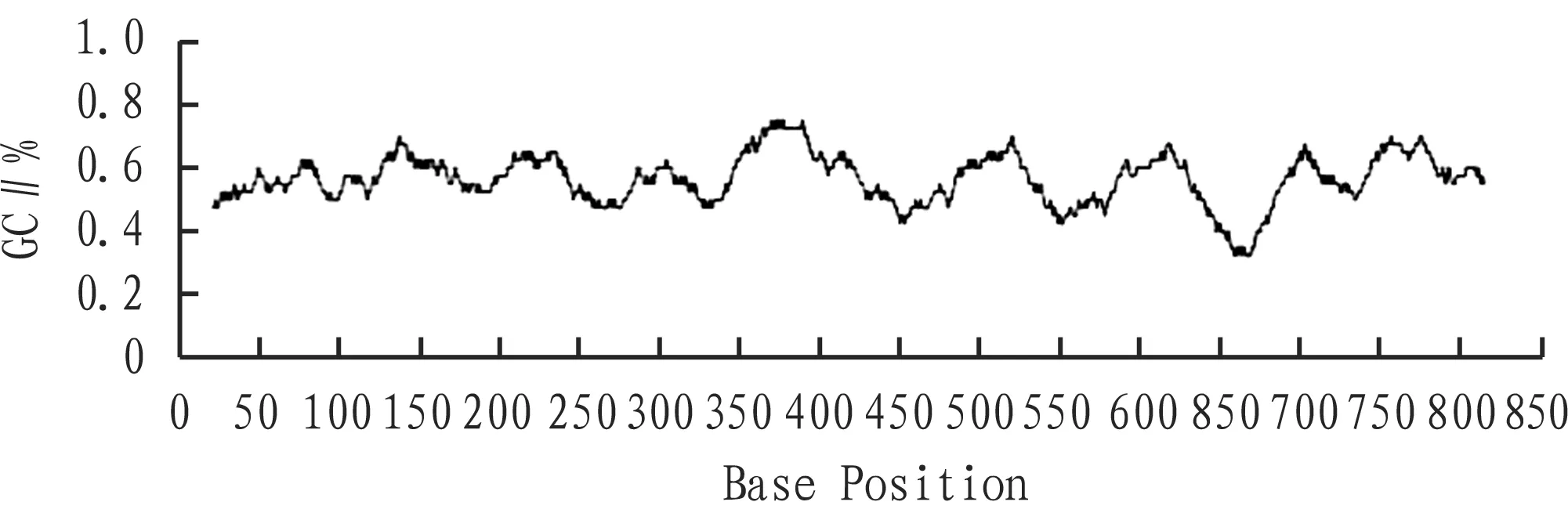
Fig.2GCcontentoftargetgene
2.1.2Target vector. Target vector was an artificially transformed Pcmv 5 carrier, and AMP resistance was added manually. The total length of vector was 19043 bp, and it was macromolecular plasmid vector (Fig. 3).
2.2Testmethods
2.2.1Synthesis of target gene. According to gene sequences, the related companies were commissioned to design and synthesize 26 primers required by PCR amplification, and the primers were diluted to 20 pmol/uL. 2 uL of solution was taken from each primer and evenly mixed as mixed primer solution, and the target gene was synthesized through two rounds of PCR (Table 1).
Fig.3Vectorstructure
Table1Primersequence

SerialnumberSequence(5to3)Basenumber1GGGGCTGCTGACCTGGCTCATGTCCATCGATGTCAAGTACCAGATCTGGAAG522GAACGAGTTGTCCGTGAAGATGACCCCGAACTTCCAGATCTGGTACTTGACATC543TCTTCACGGACAACTCGTTCCTGTACCTGGGCTGGTACATGGTGATGTCC504CGGCAAAGAAGAAGTTGTTGTAGTGGCCCAGGAGGGACATCACCATGTACCAGC545CAACAACTTCTTCTTTGCCGCCCACCTGCTGGACATCGCCATGGGGGTCAAGA536TTGTGGGTGACAGAGGAGAGGATGGTACGCAGCGTCTTGACCCCCATGGC507CTCTCCTCTGTCACCCACAATGGGAAACAGCTGGTGATGACTGTGGGCCTCCT538AAGGCCACCACAGTGTACAGGTAGACCACGACGGCCAGGAGGCCCACAGTCATC549CTGTACACTGTGGTGGCCTTCAACTTCTTCCGCAAGTTCTACAACAAGAGCGA5310GCACTTCATGTCCGGCTCGTCCTCGTCCTCGCTCTTGTTGTAGAACTTGC5011AGCCGGACATGAAGTGCGATGACATGATGACGTGCTACCTGTTCCACATGTACGT5512TCGTCCCCGATGCCTCCGCCAGCCCGGACGCCCACGTACATGTGGAACAGGTAGC5513GGCATCGGGGACGAGATCGAGGACCCAGCGGGCGATGAATACGAGCTCTAC5114AAGAAGAAGGTGATGTCGAAGACCACCCGGTAGAGCTCGTATTCATCGCC5015GTCTTCGACATCACCTTCTTCTTCTTCGTCATTGTCATCCTGCTGGCCATC5116GGAGCTCGCCGAAGGCGGCGATAATCAGACCCTGGATGATGGCCAGCAGGATGA5417CCTTCGGCGAGCTCCGAGACCAGCAGGAGCAAGTGAAGGAAGATATGGAG5018CTGCCAATCCCGCAGATGAAGCATTTGGTCTCCATATCTTCCTTCACTTGCT5219CTGCGGGATTGGCAGTGACTACTTCGATACCACGCCGCACGGCTTCGAGACCC5320CATGTAATTGGCCAGATTGTGCTCCTCTAGCGTGTGGGTCTCGAAGCCGTG5121CACAATCTGGCCAATTACATGTTCTTCTTGATGTATCTGATAAACAAGGAC5122ACTCCTGGCCCGTGTGCTCCGTCTCGTCCTTGTTTATCAGATACATCAAGAA5223CACACGGGCCAGGAGTCCTACGTCTGGAAGATGTATCAGGAGAGGTGCTGGG5224ACTGCTTGCGGAAGCAGTCGCCGGCGGGGAAGAAGTCCCAGCACCTCTCCTGATA5525CTGCTTCCGCAAGCAGTACGAGGACCAGCTGAGCTGAGAAGCTTGCATGCCTGCA5526GTCACAGGGATGCCACCCGGGATCCTCTAGAGTCGACCTGCAGGCATGCAAGCTT55
2.2.2Setting of different amount of vector. 10 μL of recombinase was put into 36 PCR tubes, respectively, and these tubes were divided into 6 groups (A, B, C, D, E, F). 6 tubes in each group were numbered A1-A6, B1-B6, C1-C6, D1-D6, E1-E6 and F1-F6, respectively, and different amount of vector was added according to Table 2 (with group A as an example). Using PCR instrument, they reacted at 50℃ and were preserved at -20℃.
Table2Ligationsystem
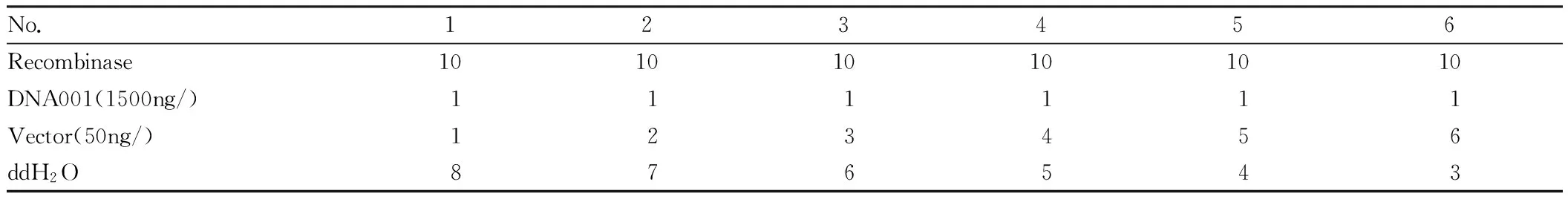
No.123456Recombinase101010101010DNA001(1500ng/)111111Vector(50ng/)123456ddH2O876543
2.2.3Setting of different competence. As shown in Table 3, different competence was set.
2.2.4Bacteria detection result judgment. There were 36 plates, and 24 plaques were picked from each plate for electrophoresis. There were too many electrophoresis samples, and the result did not directly display electrophoretogram. And the number of positive clones from every 24 clones was counted and divided by 24 to get the rate of positive clones. The positive clones of bacteria detection product were judged as follows: electrophoretic bands were matched with ladder; 1149 bp meant that the positive clones were denoted as 1; 412 bp meant that the empty vector was denoted as 0; the primer dimer meant that the variegated bacteria sample was denoted as 0.
Table3Transformationcombination

Top10FDH5αStbl3Epi400JM108SCSIA1B1C1D1E1F1A2B2C2D2E2F2A3B3C3D3E3F3A4B4C4D4E4F4A5B5C5D5E5F5A6B6C6D6E6F6
3 Results
3.1SynthesisoftargetgeneAs shown in Fig. 4, PCR products of the target gene were 750-1000 bp, consistent with the theoretical value of 855 bp.
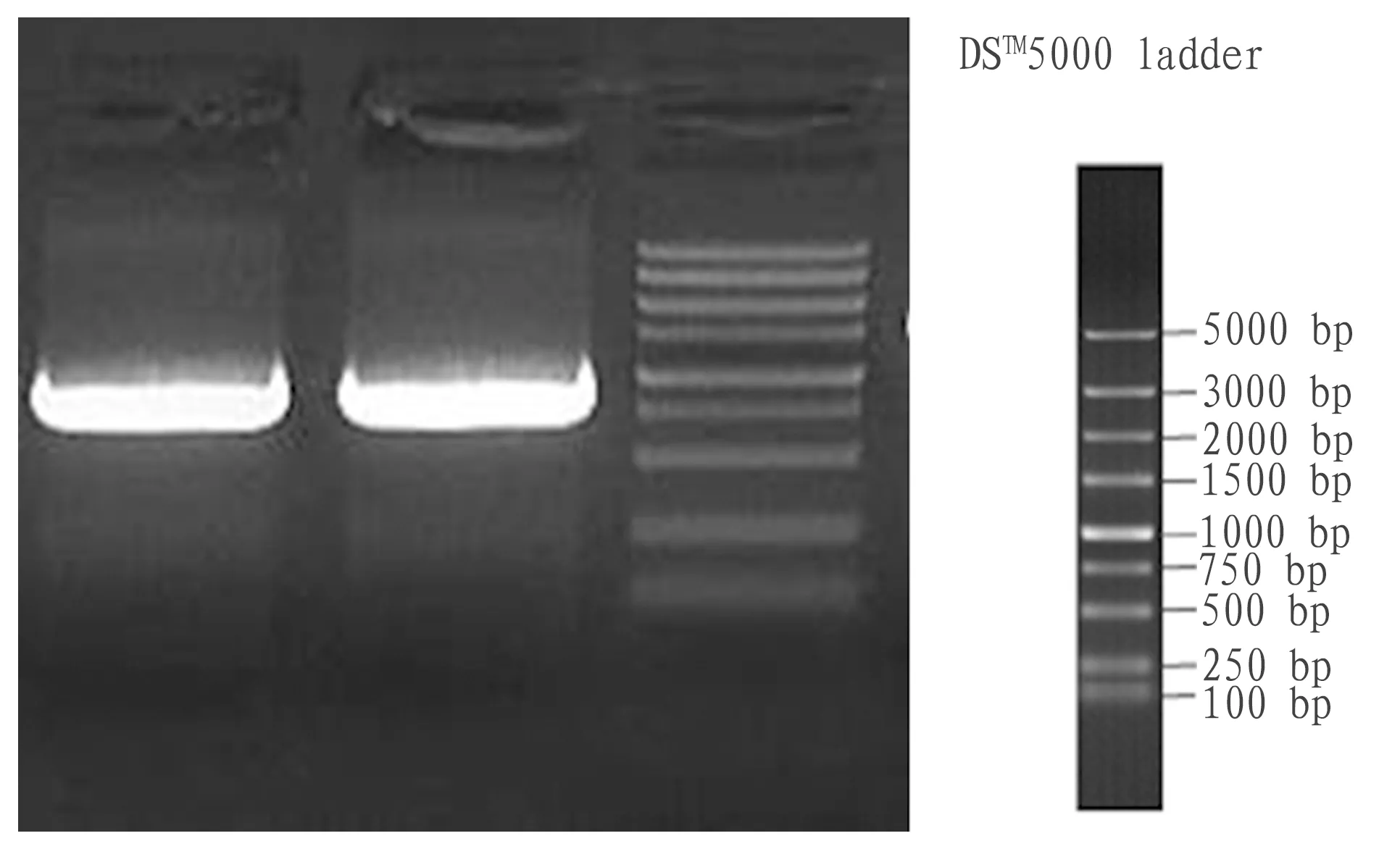
Fig.4ElectrophoretogramofPCRproducts
3.2LinearizationofvectorAs shown in Fig. 5, the theoretical size after enzyme digestion was 19043 bp and 98 bp, and the minimum value of 5000 Ladder was 100 bp, so 98 bp could not be displayed in the figure. The enzyme digestion result was consistent with the theoretical value.
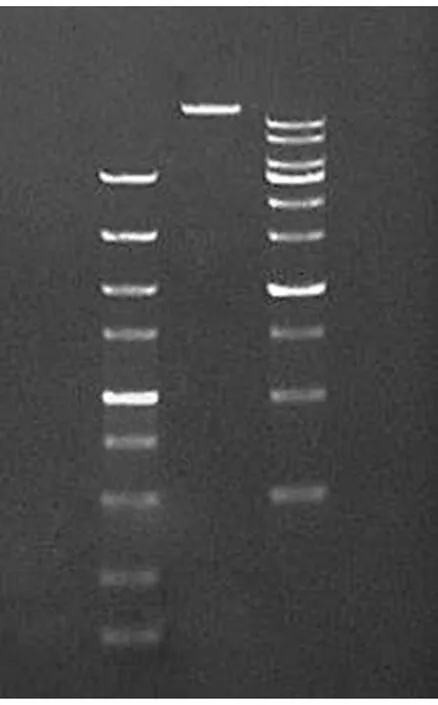
Note: Band 1, 2, 3 represented 5000 bp Ladder, enzyme digestion sample and 10 kb Ladder, respectively.
Fig.5Electrophoretogramofvectorenzymedigestion
3.3GrowthoftransformedcoloniesAs shown in Fig. 6, in 6 kinds of competence, there were a total of 36 plates (A1-F6), and the colonies with vector amount of 50, 100, 150, 200, 250 and 300 ng were added, respectively. It was clearly found that the colonies on the 36 plates grew well.

Fig.6Colonygrowthin6kindsofcompetence
3.4RateofpositiveclonesFrom Table 4 and Fig. 7, it was found that different vector density had a significant impact on the rate of positive clones of macromolecular vectors, and in terms of the average rate of positive clones, 200 ng>250 ng>300 ng>150 ng>100 ng>50 ng. In all kinds of competence, the rate of positive clones was highest at 200 ng, the highest rate was up to 75%, and the average rate reached 28.5%. If the vector density was less than 100 ng, the transformation efficiency was less than 10%. Dif-ferent competence had a significant effect on the rate of positive clones of macromolecular vector, and the average rate of positive clones was in the order of stbl3>Top10F’>DH5α>Jm108>Epi400>Scsl. The best competence was stbl3, and its rate of positive clones was higher than that of other kinds of competence under any vector density, and the average rate of positive clones was 42.4%. The average rate of positive clones was below 10% for Jm108, Epi400 and Scsl.
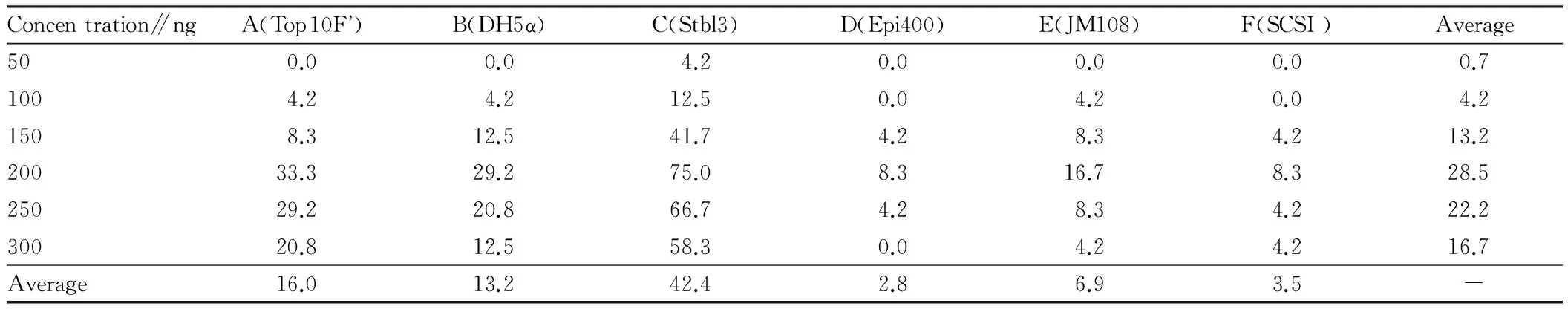
Table 4 The rate of positive clones Unit: %
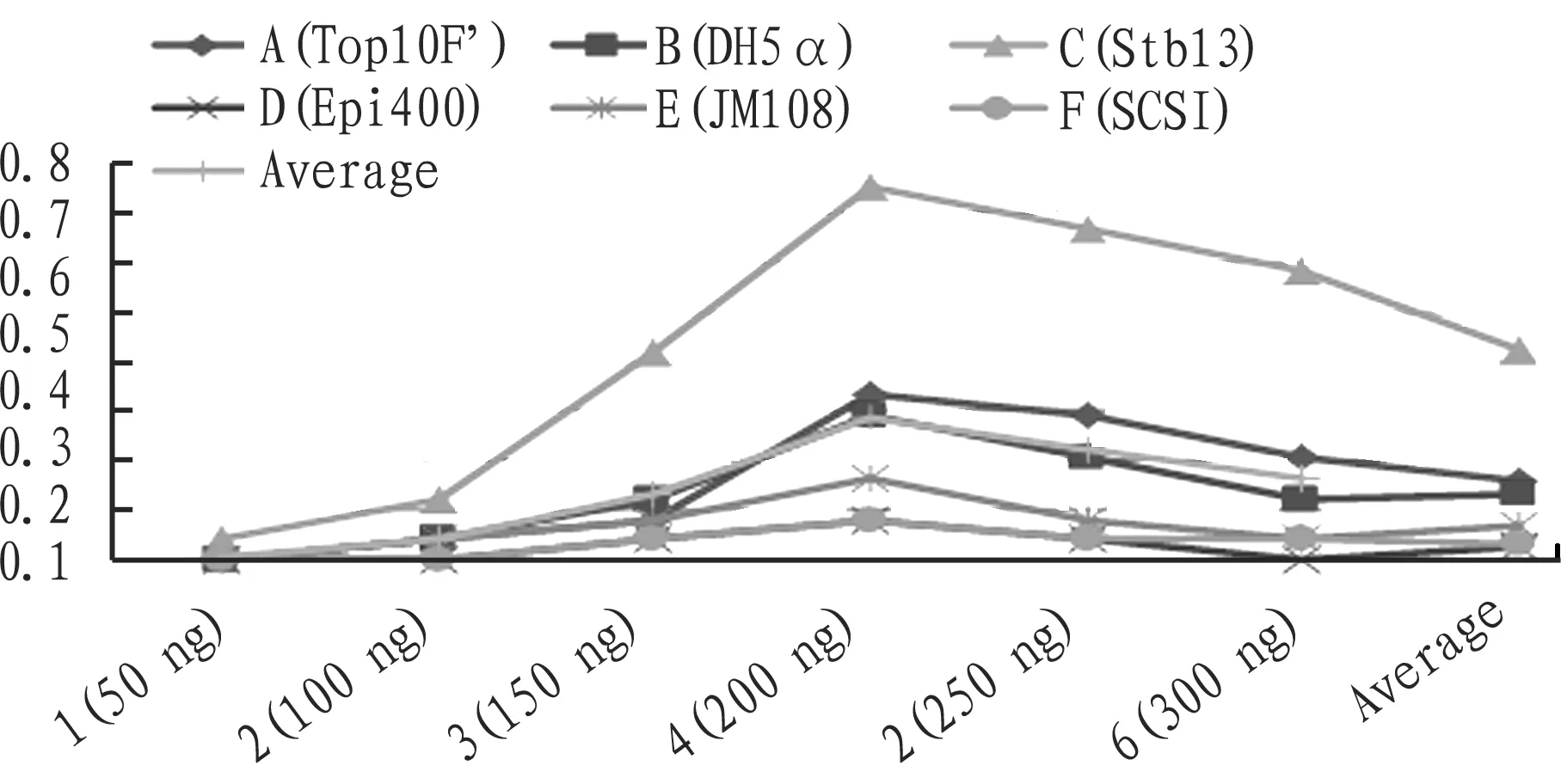
Fig.7Therateofpositiveclones
4 Conclusions and discussions
In this study, for the 20 kb macromolecular vector, on the basis of 1500 ng target gene substrate, we set the vector amount with six gradients (50 ng, 100 ng, 150 ng, 200 ng, 250 ng, 300 ng), and the recombinant reaction products obtained were transformed into six different kinds of competence (Top10F ’, DH5α, Stbl3, Epi400, JM108, SCSI) to form 36 test combinations. The results showed that both the vector amount and competence significantly affected the rate of positive clones. When the vector amount in the reaction system was too low, the recombinant reaction could not get enough positive clones, and the screening efficiency was very low. When the vector amount in the reaction system was too high, many empty vectors might be selected, affecting the rate of positive clones. For the 20 kb vector, when the vector molecular weight was about 200 ng, the rate of positive clones was highest. Compared with other kinds of competence, Stbl3 had a greater ability to copy large vectors. The optimal combination was 200 ng vector density and Stbl3, and the rate of positive clones could reach 75%. For the common recombinant test, the vector sample amount was far less than 200 ng, and taking the common puc57 series vector for example, the vector sample amount for recombination reaction was only about 30 ng[1, 3, 4]. When there was a large vector in the laboratory, the rate of positive clones might be low and even 0. Due to different genotypes for different competence, there were large differences in copying rate of different sizes and different types of plasmid[14-18]. In this study, the vector used was close to 20 kb, and it was difficult to clone compared with the commonly used 3-4 kb vector. The macromolecular vector had features of low recombination efficiency, low transformation efficiency, low copying rate, unstable expression, difficult extraction, separation and purification[8-10], so many domestic DNA companies were not willing to accept cloning of macromolecular vector, and even if there are corresponding services, the service price was high due to high failure rate and experimentation cost. In this study, with 20kb vector as example, we selected the vector amount with six gradients and six kinds of competence, indicating that the vector amount and competence indeed had a significant impact on the rate of positive clones. We obtained the optimal combination which can be used as reference for improving the cloning efficiency of macromolecular vector.
[1] ZHONG X, ZHAI C, CHEN L,etal. Construction of directional T vector for gene cloning and expression[J]. Chinese Journal of Biotechnology,2013,29( 4) : 510-519.(in Chinese).
[2] YU Y, JIANG SC, WANG KY,etal. Research progress of large fragment DNA cloning vector[J].Heilongjiang Agricultural Science,2015(2) : 147-151.(in Chinese).
[3] WU YF, LIANG DC, GUO G,etal. Construction of pUC-T vector[J].Tianjin Medical Journal,2005,33(3) : 159-160.(in Chinese).
[4] LI J, ZHANG JF, ZHEN YL,etal. Construction of recombinant plasmid pCMV-Myc-PIASx and its protein expression[J].Journal of Jilin University: Med Ed,2009,35( 3) : 415-418.(in Chinese).
[5] YAN F, ZHAO XY, DENG HX,etal. Construction and expression of a novel bisbicistronic expression vector:pCMV-Myc-IRES-EGFP[J]. Chinese Journal of Biotechnology,2007,23( 3) : 423-428.(in Chinese).
[6] WANG HZ, ZHOU XY, SONG ZX,etal. Degenerate PCR and its application in gene cloning[J].Hereditas,2003,25( 2) : 201-204.(in Chinese).
[7] ZHENG BQ, WANG Y, PENG ZH,etal. Cloning of ACC oxidase gene from Cattleya flower and construction of its plant antisense expression vector[J].Acta Agriculturae Nucleatae Sinica,2009,23( 3) : 442-446.(in Chinese).
[8] WANG H, WU WT, GU XQ. Progress in studying on liposome as an carrier for biological macromolecule[J].Pharmaceutical Biotechnology,2002,9( 3) : 171-174.(in Chinese).
[9] TAN DY, DENG SS, MENG L,etal. Improving the recombination efficiency of larger plasmid DNA by two-ligation[J].Progress in Biochemistry and Biophysics,1997,24( 3) : 281-283.(in Chinese).
[10] FAN YL, ZHAO KJ. Progress in development of large DNA fragment cloning vectors[J].China Biotechnology,2004,24( 3) : 12-16.(in Chinese).
[11] LIU L,LIU Y G,XU X P,etal. Efficient linking and transfer of multiple genes bu a multigene assembly and transformation vector system[J].PNAS,2003,100( 10) : 5962-5967.
[12] SHIZUYA H,KOUROS-MEHR H.The development and applications of the bacterial artificiakl chromosome cloning system[J]. Keio Journal of Medicine,2001,50( 1) : 26-30.
[13] ZHANG FF, SHI MD, SHANG GD. Construction of higher conversion efficiencyEscherichiacoliBL21( DE3) via recombineerin[J].Journal of Anhui Agricultural Sciences,2015,43( 20) :29-31,72.(in Chinese).
[14] ZHANG LL, XU CY, XU CJ. Factors affecting transformation ability of competentEscherichiacolicells[J].Chinese Journal of Cell Biology,2004,26( 4) : 429-432.(in Chinese).
[15] YANG K, GONG ZH, LI DW. The preparation of super E.coli competent cell and the establishment of fast plasmid transformation intoE.coli[J].Northern Horticulture,2010( 14) : 127-130.(in Chinese).
[16] DAI J.Optimization on the preparation of the competent cells ofEscherichiacoli[J]. Jiangsu Agricultural Sciences,2015,43( 4) : 53-54.(in Chinese).
[17] YANG K. The establishment of competence cell transformation system and the construction of transient expression vector[D].Xianyang: North West Agriculture and Forestry University,2010: 1-48.(in Chinese).
[18] TU ZM, CHEN MJ, HE GY,etal. Competent cells preparation and plasmid transformation for threeEscherichiacolistrains[J].Journal of Huazhong University of Science and Technology(Nature Science Edition),2006,34( 4) :112-115.(in Chinese).
 Asian Agricultural Research2016年12期
Asian Agricultural Research2016年12期
- Asian Agricultural Research的其它文章
- How to Improve Tea Farmers’ Livelihoods in the Sightseeing Place along Lijiang River Valley?
- Brand Construction of Agricultural Enterprises: A Case Study of Hubei Hanway Ecological Agriculture Group
- Impact of Online Comments on Purchase Intention of College Student Consumers under Online Shopping
- Empirical Research on Farmer’s Breaching Behavior in Order Contract
- Suitability Evaluation of Garden Landscape in Chizhou Residential Area
- Evaluation on the Level of Agricultural Informatization Development in Gansu Province during 2001-2010
