镍、镉与去甲基斑蝥酸钠和咪唑配合物的结构、与DNA/BSA的作用及抗增殖活性
杜芳园 李士坤 林秋月*, 魏 琼 汤宁宁
(1浙江师范大学浙江省固体表面反应化学重点实验室,金华321004)
(2浙江师范大学化学与生命科学学院,金华321004)
杜芳园1,2李士坤2林秋月*,1,2魏 琼2汤宁宁2
(1浙江师范大学浙江省固体表面反应化学重点实验室,金华321004)
(2浙江师范大学化学与生命科学学院,金华321004)
以2种配体去甲基斑蝥酸钠(Na2DCA=7-氧杂二环[2.2.1]庚烷-2,3-二甲酸钠)和咪唑(IM),分别与镍和镉的醋酸盐通过溶液法合成了2种配合物[Ni(IM)(DCA)(H2O)2]·2H2O(1),[Cd2(IM)4(DCA)2]·2H2O(2)。应用元素分析、热重分析、红外光谱及X-射线单晶衍射法对配合物的组成和结构进行了表征。配合物1与2的中心离子分别与咪唑的亚胺氮原子、去甲基斑蝥酸根的羧基氧原子和醚键氧原子配位,配位数均为6,分别为单核(1)和双核(2)配合物。通过紫外光谱法、荧光光谱法和粘度法研究了配合物与DNA的相互作用。结果表明,配合物能通过部分插入模式与DNA发生较强的结合作用(Kb:5.51×103(1)、1.01×103(2)L· mol-1)。同时,利用荧光光谱法研究了配合物与牛血清白蛋白(BSA)的相互作用。配合物能与BSA发生强烈的相互作用(KA:1.91× 105(1)和6.17×105(2)L·mol-1),结合位点数均为1。测试了配合物对人肝癌细胞(SMMC7721)和人乳腺癌(MCF-7)的体外抗增殖活性。结果显示,配合物对不同癌细胞具有选择性抑制作用。镍配合物(1)对SMMC7721的抗癌活性较去甲基斑蝥酸钠有明显提高。
咪唑;去甲基斑蝥酸钠;镍配合物;镉配合物;与DNA和BSA相互作用;体外抗增殖活性
0 Introduction
Recent studies indicated that the transition metal complexes possessed excellent antiproliferative activities,especially complexes containing natural anticancer products[1-2].Disodium demethylcantharate(Na2(DCA)), as the derivatives of cantharidin,have been applied in clinical use[3].Some previous researches also showed that,after forming metal complexes,the antiproliferative activities of ligand could be improved significantly[4-5].Meanwhile,demethylcantharate(DCA)itself could inhibit the activities of PP1 and PP2A effectively[6-7].According to the literature[8],the platinum complexes containing DCA are likely to possess dual anti-cancer mechanism:inhibition of PP2A and platination of DNA,which promotes the anticancer activities of complexes.
The imidazole group is an important topic in biochemistry.It presents in the metallic centers of metalloproteins and metalloenzymes.In the active centers of biomolecules,the nitrogen atom from imidazoles is known to bond to metals.Developing new medicines that are able to treat severe poisoning is also relevant to the study of heteroligand complexes[9]. And the transition metal complexes of demethylcantharidin and heterocyclic compounds possessed intense antiproliferative activities[10-11].The interaction mechanisms of these complexes with biomacromolecules have been studied previously[12-13].
Thus,designing and synthesizing the drugs, which could simultaneously interacting with various biomacromolecules(such as DNA and protein) strongly,has a vital significance.In this study,two Niand Cdcomplexes of demethylcantharate and imidazole were designed and synthesized.The interactions of the complexes with DNA and bovine serum albumin(BSA)were investigated.In addition, antiproliferative activities against human hepatoma cells(SMMC-7721)and human breast cancer cells (MCF-7)lines were tested in vitro.
1 Experimental
1.1 M aterials and instrum ents
All reagents and chemicals were obtained from commercial sources.Demethylcantharidin(NCTD, C8H8O4)was obtained from Nanjing Zelang Medical Technology Co.Ltd.;imidazole(IM,C3H4N2)and DNA were obtained from Sinopharm Chemical Reagent Co. Ltd.;ethidium bromide(EB)was obtained from Fluka Co.Ltd.;DNA solution(ρ=200 μg·m L-1,c=3.72×10-4mol·L-1,A260/A280=1.8~2.0)was prepared by 50 mmol· L-1NaCl;BSA was purchased from Beijing BioDee BioTech Co.Ltd.and was stored at 277 K;BSA solution(ρ=500 μg·mL-1,c=7.47 μmol·L-1)was prepared by 5 mmol·L-1NaCl;Human hepatoma cells (SMMC7721)and human breast cancer cells(MCF-7) were purchased from Shanghai Institute of Cell Bank. Na2DCA was synthesized by Demethylcantharidin (NCTD)and NaOH according to the reference[14].Other chemical reagents in analytical reagent grade were used without further purification.
Elemental analyses of C,H and N were carriedout in Vario ELⅢelemental analyzer.Infrared spectra were measured using the KBr disc method by NEXUS-670 FT-IR spectrometer in the spectral range 4 000~400 cm-1.The thermogravimetric analyses were monitored on TGA/SDTA851ethermo gravimetric analyzer.Diffraction intensities of the complexes were collected at 293 K on Bruker SMART APEXⅡCCD difffractometer.Electronic absorption spectra were recorded on UV-2501 PC spectrophotometer.Fluorescence emission spectra were obtained by Perkin-Elmer LS-55 spectrofluorometer.Viscosity experiments were carried on Ubbelodhe viscometer.
1.2 Syntheses of the complexes
A mixture of Ni(Ac)2·4H2O(1 mmol)and imidazole(1 mmol)were dissolved in water.The aqueous solution of Na2DCA(1 mmol)was added dropwise to the mixed solution.The green solution was then filtered after stirring for 4 h.Two weeks later,green crystals with suitable size for single-crystal X-ray diffraction were obtained.Anal.Calcd.(%)for NiC11H20N2O9:C,34.50;H,5.26;N,7.31.Found(%):C,34.35; H,5.32;N,7.40.Λm(DMF):21 S·cm2·mol-1.IR spectra (KBr,cm-1):1 604(νas(COO-));1 416(νs(COO-));1 267, 1 185,988(ν(C-O-C));737,584(ν(O-H)).
The solution of Cd(Ac)2·4H2O(1 mmol)and imidazole(3 mmol)were stirred at 50℃for 2 h.The aqueous solution of Na2DCA(1 mmol)was added dropwise to the mixed solution.And the pH value of the mixed solution was adjusted to 5.0 using glacial acetic acid.The colorless solution was then filtered after stirring at 50℃for 4 h.Two weeks later, colorless crystals with suitable size for single-crystal X-ray diffraction were obtained.Anal.Calcd.(%)for Cd2C28H36N8O12:C,37.31;H,4.02;N,12.43.Found (%):C,37.48;H,3.95;N,12.26.Λm(DMF):25 S·cm2·mol-1.IR spectra(KBr,cm-1):1 595(νas(COO-));1 391 (νs(COO-));1 265,1 178,987(ν(C-O-C)).
1.3 Antiproliferative activity evaluation
The MTT assay was applied to measure the antiproliferative activities.Title complex was dissolved in DMSO as 100 mmol·L-1stock solutions and then diluted in culture medium before using.The final concentration of DMSO in the medium was less than 0.1%and no interference with the tested biological activities was shown[15].Cells were seeded for 24 h, and the title complex or Na2(DCA)was added and incubated for 72 h.Then 100 μL MTT(1 mg·m L-1, dissolved in culture medium)was added into each well and incubated for 4 h(37℃).The inhibition ratio was calculated.The errors quoted were standard deviations,which were based on three replicates[16].
1.4 Crystal structure determ ination
Single crystals,with size of 0.310 mm×0.163 mm×0.036 mm(1)or 0.199 mm×0.179 mm×0.138 mm (2),were analyzed by X-ray diffraction.Data were collected with a graphite monochromatic Mo Kα radiation(λ=0.071 073 nm)using the ω-2θ scan technique at 296(2)K.The structures were solved by direct methods and refined by full-matrix least-squares techniques using the SHELXTL-97 program package[17-18]. All non-hydrogen atoms were refined anisotropically. Besides the hydrogen atoms next to oxygen atoms located on different Fourier maps,other hydrogen atoms were generated geometrically.Crystal data and experimental details for structural analyses are listed in Table 1.
CCDC:822330,1;822328,2.
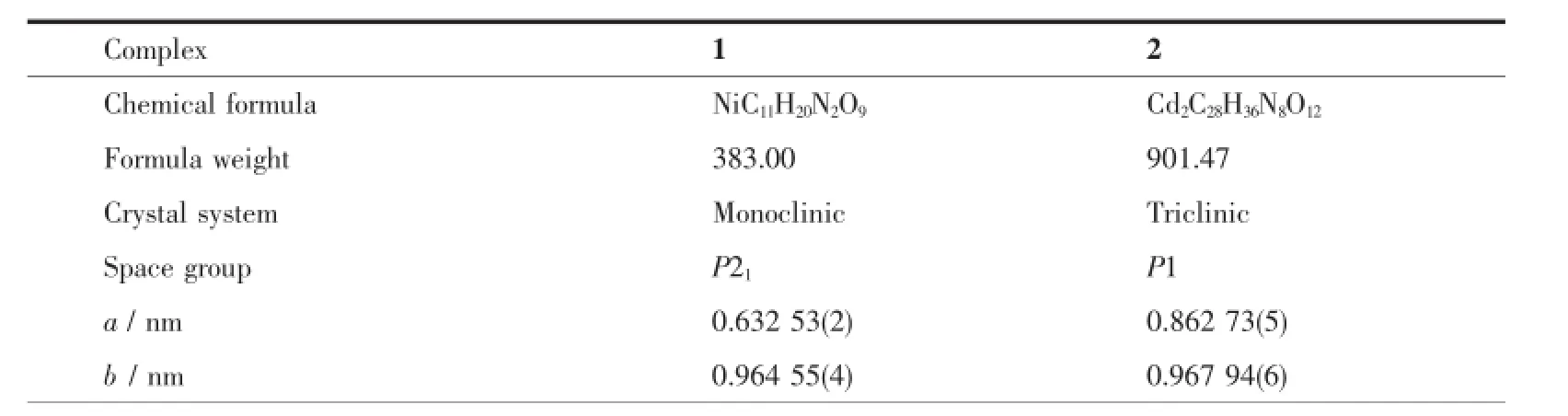
Table1 Crystal data and structure refinem ent details for the com p lexes
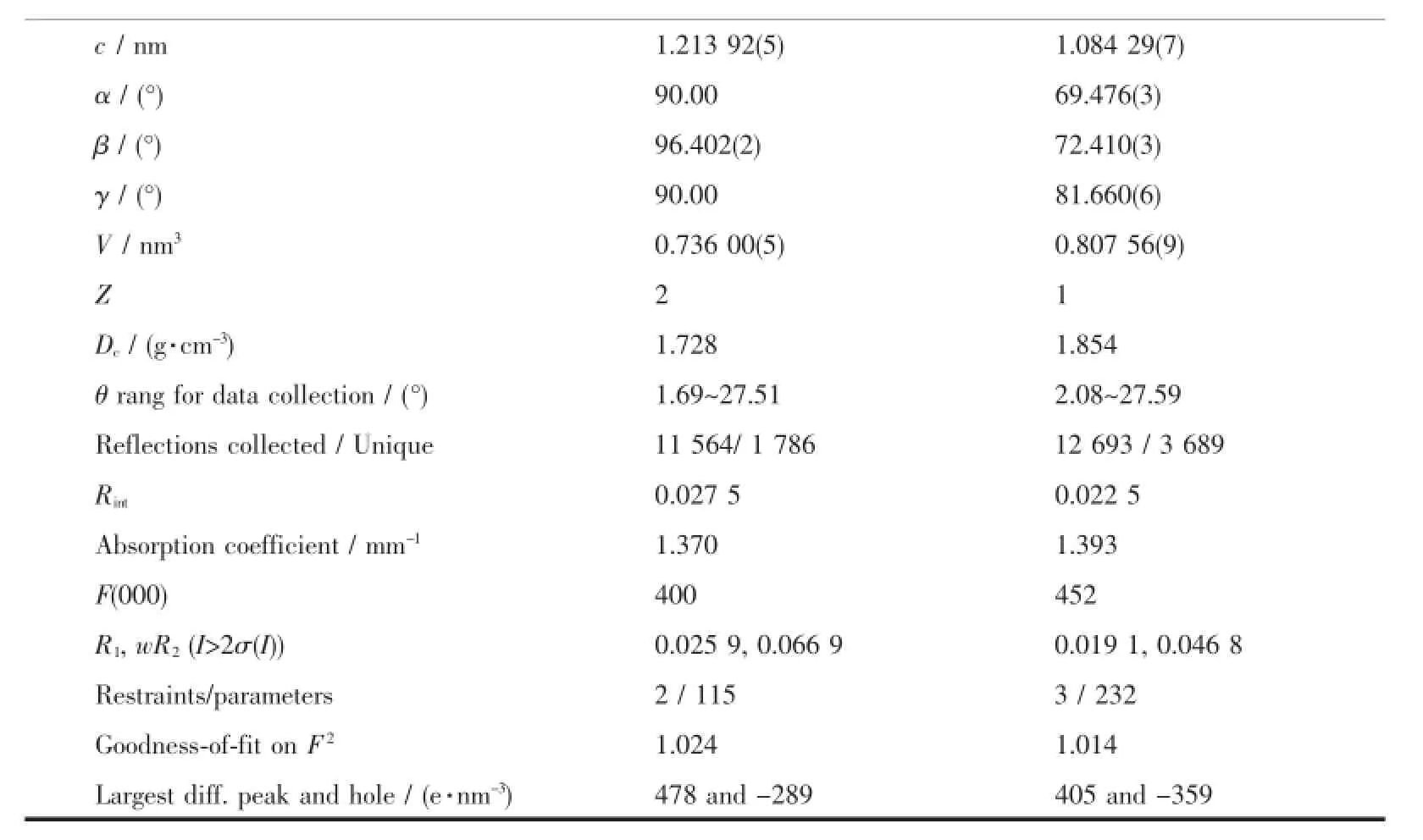
Continued Table 1
2 Results and discussion
2.1 Characterization and crystal structure
2.1.1 Thermogravimetric analysis(TGA)
The experiment was performed under air atmosphere with a heating rate of 10℃·min-1and temperature range of 30~800℃.The TG-DTG curves of complex 1 are shown in Fig.1.For complex 1,according to the TG-DTG curves,a four-stage weight loss processes happened.The first weight loss stage(9.98%) occurred in the temperature range of 65~160℃, which corresponded to the departure of the two crystal water(9.40%theoretical).The second weight loss stage(9.48%),which corresponded to the departure of two coordinated water(9.40%theoretical),was occurred in the temperature range of 161~215℃.The third weight loss stage(16.86%),which corresponded to the departure of imidazole ligand(17.78%theoretical), was occurred in the temperature range of 216~340℃. Title complex gave a sharp weight loss peak at temperature around 341~525℃,which corresponds to the weight loss of 43.96%.It indicates the thermal decomposition of one DCA(43.92%theoretical).At temperature above 525℃,no further weight loss occurred.The sample residue was NiO,which weigh 19.72%of the initial mass(19.50%theoretical).The TG-DTG curves of complex 2 were similar to 1.
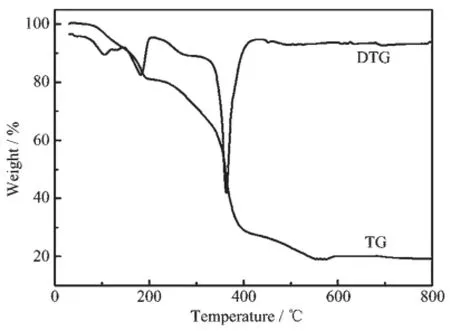
Fig.1 TG-DTG curves of complex 1
2.1.2 Structural description of the complexes
Molecular structure of complexes were shown in Fig.2.Selected bond lengths and bond angles are presented in Table 2 and 3.
Molecular structure of complex[Ni(IM)(DCA) (H2O)2]·2H2O(1)is shown in Fig.2a.The structural model shows the Niion was six-coordinated.Each Niwas coordinated with one imine nitrogen N(1) from imidazole(IM),two carboxylate oxygen atoms O(1) and O(1A)in two different carboxylate groups,one bridge oxygen atom O(3)from demethylcantharate,and two oxygen atoms(O(1W#1),O(1W),)from water.The bond angles of O(1)#1-Ni(1)-O(1),O(1W)#1-Ni(1)-O(1W),O(1W)-Ni(1)-O(1)and O(1W)#1-Ni(1)-O(1)#1 are 84.57(7)°,88.74(8)°,176.81(5)°and 176.81(5)°,respectively,all of which are close to 90°.Thus,a slightly distorted quadrangle was formed around Ni(1)by O(1W),O(1W)#1,O(1),and O(1)#1.Similarly,the imine nitrogen N(1)and the bridge oxygen atom O(3) were in the axial positions.They formed a distorted octahedral structure.Due to the binding of the bridge oxygen atoms O(3)with Ni,two six-membered rings (Ni(1)-O(3)-C(3)#1-C(2)#1-C(1)#1-O(1)#1 and Ni(1)-O(3)-C(3)-C(2)-C(1)-O(1))were formed,and a sevenmembered ring(Ni(1)-O(1)-C(1)-C(2)-C(2)#1-C(1)#1-O(1)#1)was formed,which could have stabilized the complex.
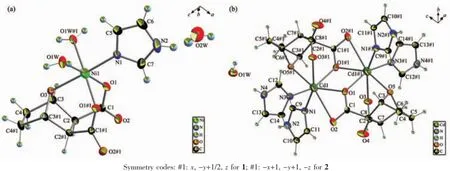
Fig.2 Coordination environment of metal atoms in the complex 1(a)and complex 2(b)showing 50%probability ellipsoids and the atom-labeling scheme
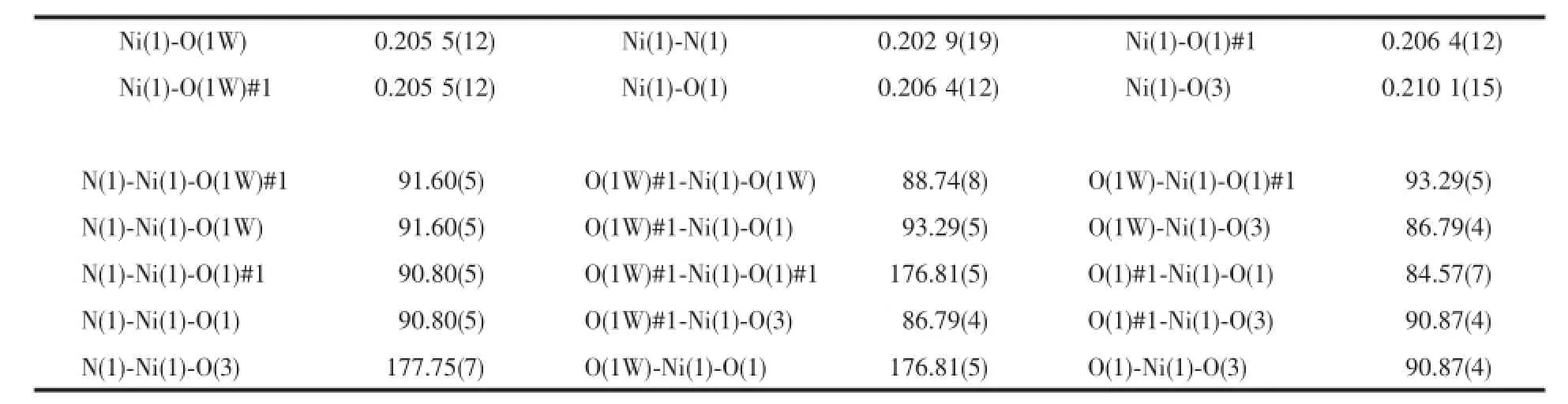
Table2 Selected bond lengths(nm)and angles(°)for complex 1
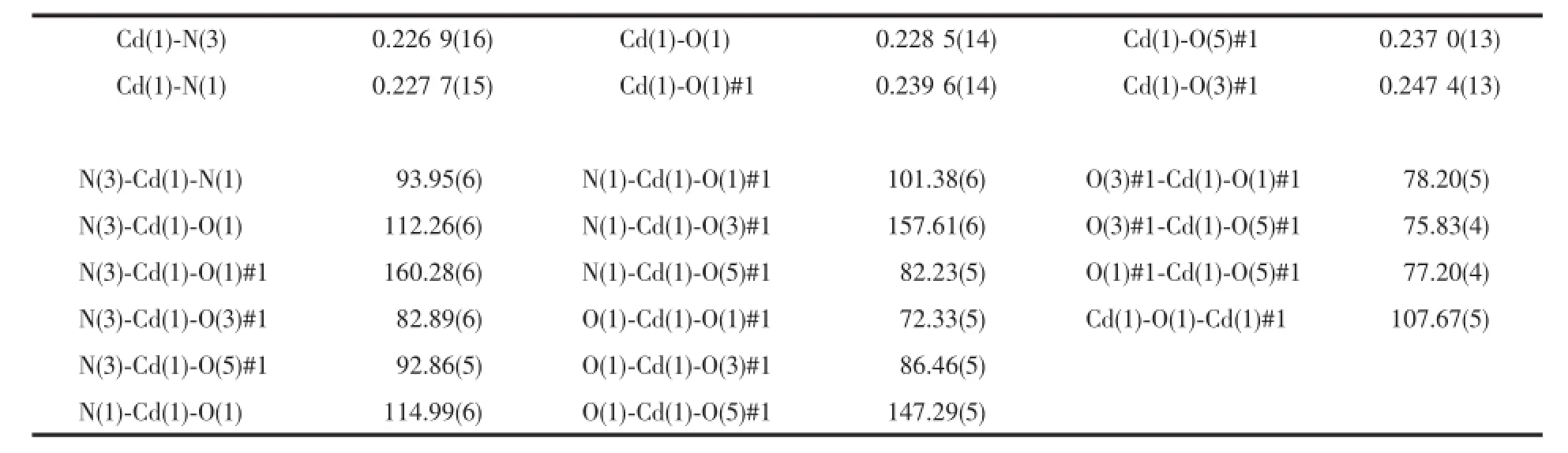
Table3 Selected bond lengths(nm)and angles(°)for com p lex 2
Molecular structure of complex[Cd2(IM)4(DCA)2]· 2H2O(2)is shown in Fig.2b.The complex was binuclear,and each Cdion was six-coordinated.Each Cdion was six-coordinated by two nitrogen atoms from two imidazole,one ether oxygen atom,two oxygen atoms of different carboxyl groups from one DCA,and one bridging oxygen atom of carboxyl group from the other DCA.Two Cdions were connected by O(1)andO(1)#1,and formed a quadrangle coordination center Cd2O2,which was the symmetric center of the entire molecule.O(1),O(1)#1,O(5)#1 and N(3)lay on the same equatorial plane with Cd(1)with a torsion angle. O(3)#1 and N(1)were in the axial positions.It formed a distorted octahedral structure since the bond angle of N(1)-Cd(1)-O(3)#1 was 157.61(6)°.And two imidazole molecules that coordinated with each Cdion were vertical to each other.
Packing diagram of the complexes was showed in Fig.3,and the hydrogen bond lengths and bond angles are given in Table 4 and 5.
Taking complex 1 as an example,strong(O-H…O)hydrogenbonds(O…O distance is0.27480~0.28696 nm)existed between adjacent chains,which involve the coordinated water,the crystal water and the carboxyl group from adjacent chains.Numerous intraand intermolecular hydrogen-bonding interactions were formed.Crystal water of each molecular was linked with O(2W)-H(2WA)…O(2W)#4,resulting in an onedimensional chain.These one-dimensional chains could extend the structure to 2D layer with O(1W)-H(1WA)…O(2)#2 and O(2W)-H(2WB)…O(2)#1.And these layers could constructed the three-dimensionalstructure of the complex with O(1W)-H(1WB)…O(1)#3. Therefore,we concluded that the synergistic effect, such as hydrogen-bonding interactions,existing between the complexes and biomacromolecule,could be the fundamental cause of the biological activity change found in macromolecules.
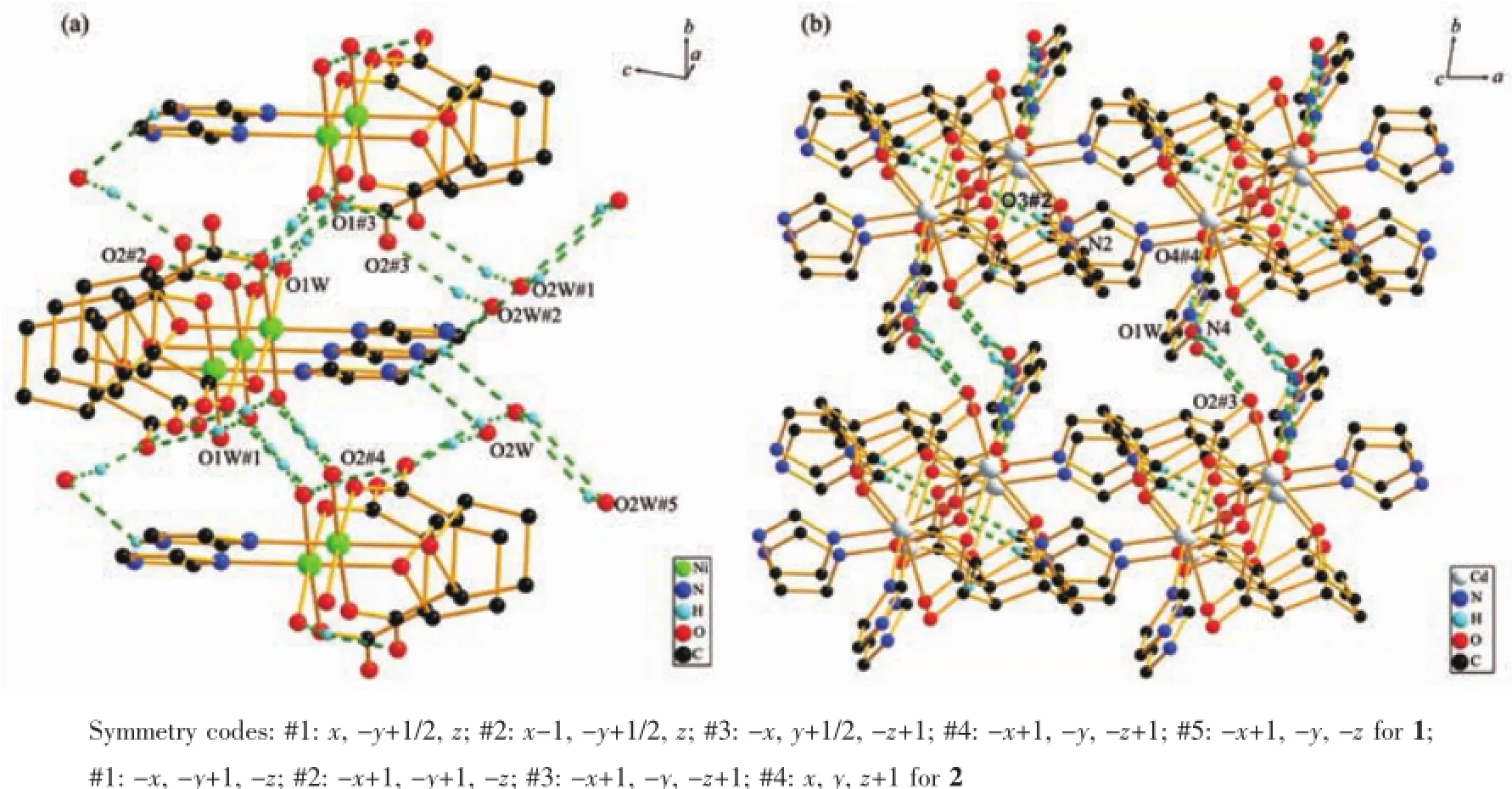
Fig.3 Packing diagrams of complex 1(a)and complex 2(b)showing hydrogen bonding interactions(dashed lines)
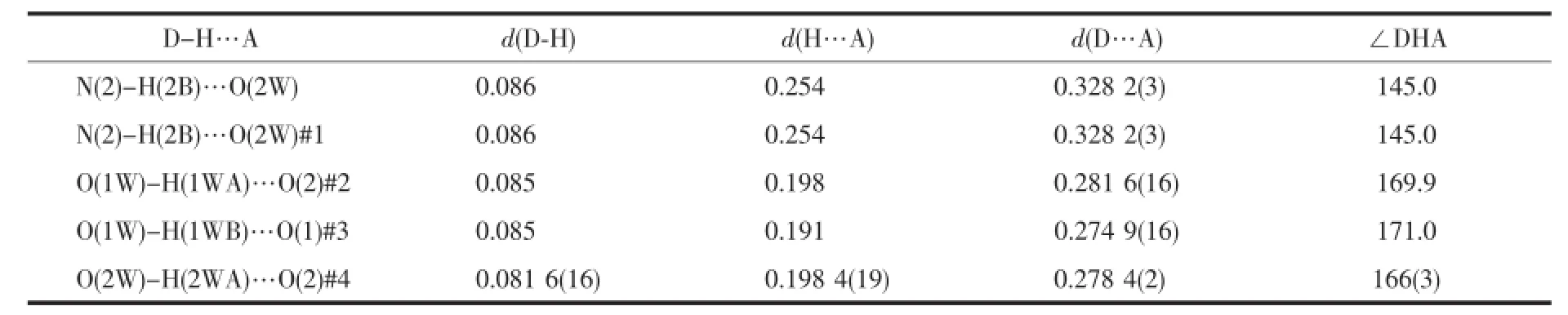
Table4 Hydrogen bond lengths(nm)and bond angles(°)for com plex 1

Table5 Hydrogen bond lengths(nm)and bond angles(°)for com plex 2
2.2 DNA binding studies
2.2.1 Electronic absorption spectra
The application of electronic absorption spectroscopy is one of the most useful techniques in DNA-binding studies[19].The UV absorption spectra would change in accordance with the environmental condition changes,since the stacking interactions happened between the complexes and DNA[20].Results are shown in Fig.4,with increasing DNA concentration, hyperchromic(1)or hypochromic(2)effect was observed,which indicated molecular level interactions existed between the complexes and DNA.
The intrinsic binding constant(Kb)was determined by the equation:cDNA/(εA-εF)=cDNA/(εB-εF)+1/[Kb(εB-εF)], where εA,εFand εBcorresponded to the apparent extinction coefficient,the extinction coefficient for the free compounds and its fully DNA-bound combination, respectively[21].Kbvalues of the complexes were 3.28× 104(1),5.95×103(2)L·mol-1,respectively.These values suggested that the complexes possessed moderate binding abilities with DNA.Meanwhile,the binding constants of the complexes with DNA were one or two orders of magnitude less than that of classical intercalator(EB)[22],which indicated the binding mode between title complexes and DNA were non-classical intercalative.The values suggested that the binding ability of complex 1 was more intense than complex 2, and the Kbof complex 1 is close to the value of cobaltcomplex[Co(IM)(DCA)(H2O)2]·2H2O(2.62×104L· mol-1)[23].The results suggested that the structure of the complex played an important role in the interaction with DNA.
2.2.2 Fluorescence spectral studies
To study the intensity and mode of the interaction between complexes and DNA,EB was used as fluorescence probe due to its conjugate rigid plane structure.And the fluorescence quenching of EB-DNA system by this planar structure compound was studied.Intense fluorescence was observed at 592 nm in the EB-DNA system,but not in the complexes. The results were shown in Fig.5.With increasing concentration of the complexes,the intensity of emission bands of EB-DNA system reduced,which inferred that two complexes bonded to DNA.These complexes could replace EB from EB-DNA system, and inserted into DNA string.
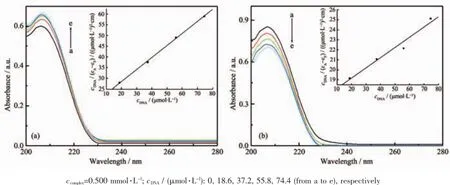
Fig.4 Absorption spectra of complex 1(a),2(b)in the absence and the presence of DNA
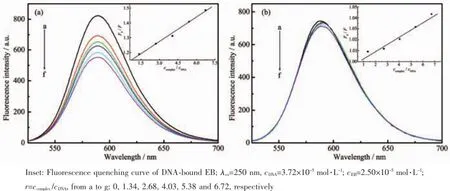
Fig.5 Emission spectra of EB-DNA system in the absence and presence of complex 1 and complex 2
According to the Stern-Volmer equation[24]:F0/F= 1+Ksqr,the abilities of complexes binding with DNA could be quantified.From the illustration in Fig.5,a preferable linear relation existed between F0/F and r, and the constant Ksqof Stern-Volmer was gradient of the illustration.The Ksqvalues for complexes were 0.055(1)and 0.006(2),respectively.The results indicated the DNA binding ability of complex 1 was more intense than complex 2,and the binding mode between the complexes and DNA was intercalation.However, the intercalation between complex 2 and DNA was too weak for complex 2 to displace EB from the EB-DNA system,and the fluorescence system has no significant change.It was attributed to the differences of their structures.Space steric hindrance resulting from two mutually perpendicular imidazole rings,hindered the insertion of complex 2 into the base pair pleated sheet of the DNA double helix.
2.2.3 Viscosity measurements
Hydrodynamic measurements are sensitive to DNA length change and considered to be the most critical tests in evaluating binding modes in solution in the absence of crystallographic structural data[25]. The classical intercalative mode can lead to significant increase in viscosity of DNA solution,due to the increasing of overall DNA length caused by separation of base pairs at intercalation sites.In contrast,a partial or non-classical intercalation of the compounds can bend or kink DNA helix,resulting in a decrease of its effective length and viscosity[26].To further study the binding mode of the compounds interacting with DNA,DNA viscosity at 25℃was investigated(Fig.6),and the relative viscosities η were calculated through equation[27]:η=(t-t0)/t0,where t0and t represent the flow time of DNA solution in the absence and presence of complex through the capillary, respectively.The experimental data showed that the relative viscosity of DNA steadily decreased after adding complexes,but there were no significant viscosity change occurred after adding Na2DCA or IM. A possible explanation is that the complexes were partially inserted to the DNA base pairs and resulting in a kink in the DNA helix,therefore the DNA effective length was reduced[28].The steric hindrance of the complex was accrescent due to the non-planar structure of demethylcantharate(DCA).
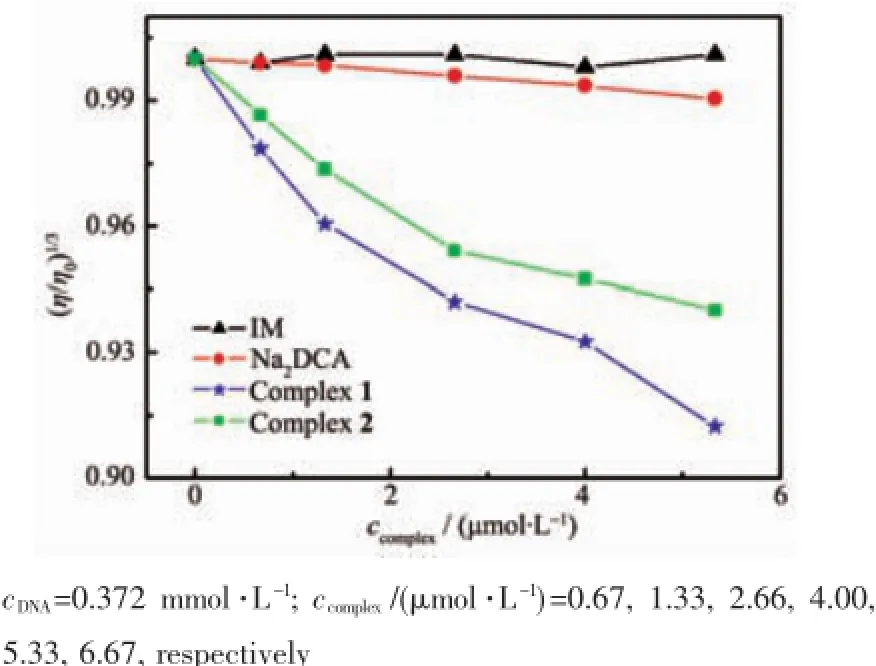
Fig.6 Effect of increasing amounts of the compounds on the relative viscosity of DNA at 25℃
Meanwhile,the relative DNA viscosity was reduced in different degrees,which inferred that complex 1 had stronger interaction with DNA than complex 2.This result agrees with the electronic absorption spectra and fluorescence spectra observation. These conclusions explained the structural differences causing the differences in DNA bindings.
2.3 Interaction w ith BSA
2.3.1 Fluorescence spectra and quenching mechanism
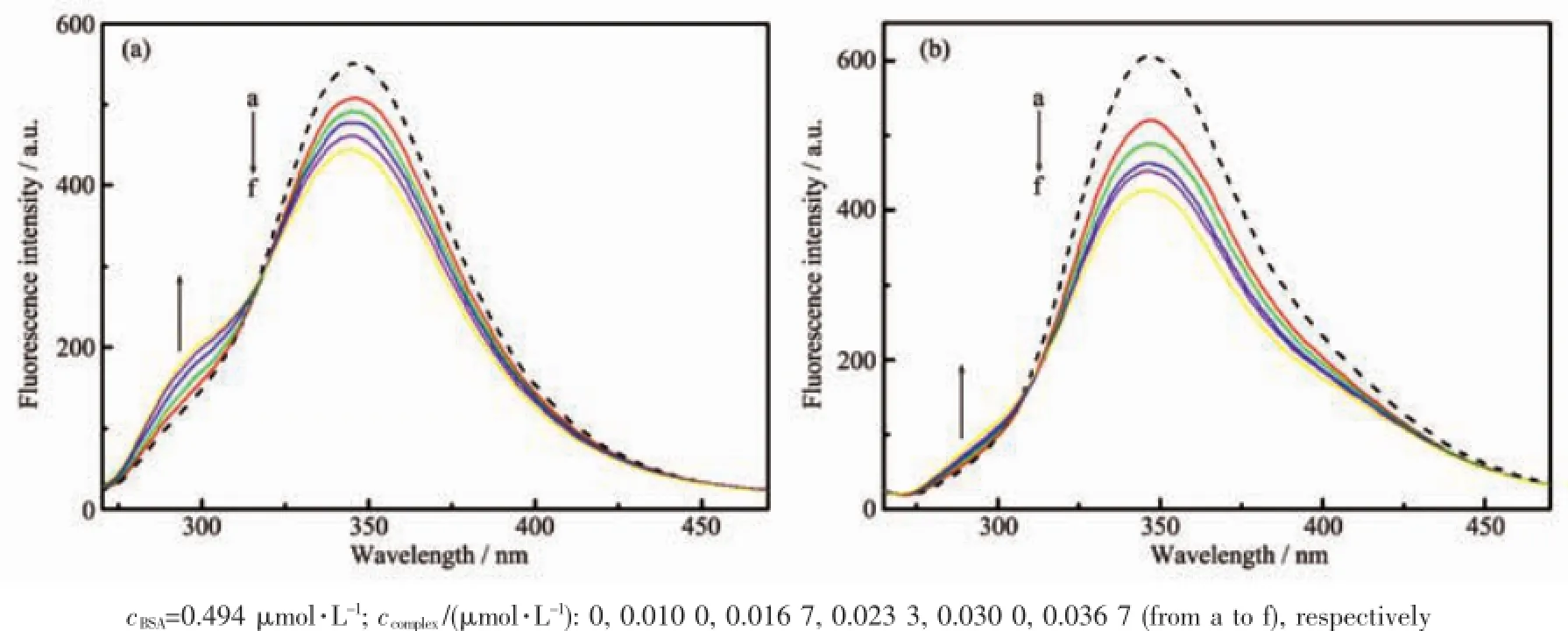
Fig.7 Fluorescence spectra of BSA in the absence(dash line)and the presence(solid line)of complex 1(a)and complex 2(b)
The fluorescence quenching of BSA by the complexes was showed in Fig.7.The results showed that BSA has strong fluorescence emission at 346 nm. The peak intensity decreased with the increasing concentration of complexes,which inferred that strong interactions and energy transfer between complexes and BSA existed[29].The BSA-complex compound system appeared one equal strength emission point at 316(1) and 310(2),indicating that a compound system was formed between BSA and complexes,resulting quenching in the BSA fluorescence[30].
In order to verify the quenching mechanism,the fluorescence quenching was assumed to be dynamic quenching.The quenching rate constants can be calculated by the Stern-Volmer equation[31]:F0/F=1+ Kqτ0cQ,where cQis the concentration of the complexes. For many proteins,τ0is known to be approximately equal to 10-8s.Using these assumptions,the calculated quenching rate constants Kqwere 5.64×1014(1)and 9.20×1014L·mol-1·s-1(2),respectively.These values were much greater than the maximum possible value for diffusion-limited quenching in water(2×1010L· mol-1·s-1),which suggested that quenching mechanism of complexes to BSA was static quenching[32].
2.3.2 Binding constants and binding sites
Assuming there were n identical and independent binding sites in protein,the binding constant KAcan be calculated using equation[33]:lg(F0-F)/F=lg KA+n lg cQ. The obtained values of KAwere 1.91×105L·mol-1(1), 6.17×105L·mol-1(2),and 2.78×104L·mol-1(Na2DCA). The values of n were 0.80(1),0.82(2)and 0.66 (Na2DCA),respectively.These results indicated that strong binding interactions existed between the complexes and BSA.And the results also inferred that complex 2 had stronger interaction with BSA than complex 1,and the binding site of complexes was one. The binding intensity of complexes with BSA was stronger than with DNA,due to the binding intensity of Na2DCA with BSA was found stronger than with DNA.
2.4 Antiproliferative activity evaluation
The antiproliferative activities of Na2(DCA)and complex 1 were evaluated by human hepatoma cells (SMMC-7721)and human breast cancer cells(MCF-7) using the MTT assay in vitro.The relations between inhibition rates and complex concentrations against human hepatoma cells and breast cancer cells were shown in Fig.8 and Fig.9,respectively.
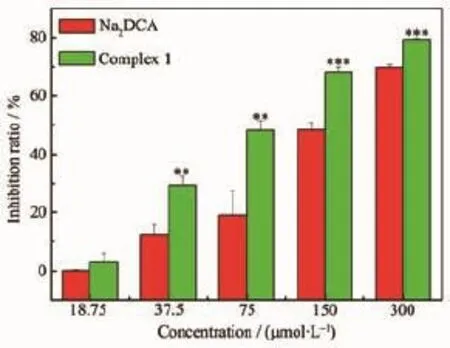
Fig.8 Inhibition effects of complex and Na2(DCA)on SMMC-7721 cell growth
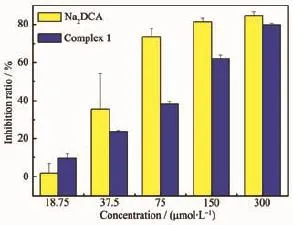
Fig.9 Inhibition effects of complex and Na2(DCA)on MCF-7 cell growth
As shown in Fig.8,the inhibition ratios of complex against human hepatoma cells were significantly higher than Na2(DCA).Especially at the concentration of 75.00 μmol·L-1,the inhibition ratio of complex ((48.2±3.4)%)was much greater than Na2(DCA) ((19.1±8.3)%).The inhibition ratios of complex 1 against SMMC-7721 lines increased with concentration growth.The concentration of the compounds with 50% inhibition(IC50)on the SMMC7721 was determined. The values of IC50of complex((86.48±6.1)μmol·L-1) and Na2(DCA)((152.8±15.6)μmol·L-1)inferred that the complex had stronger inhibition activities against human hepatoma cells compared to Na2(DCA),and it was even more intense than NCTD(IC50=(115.5±9.5) μmol·L-1)[34],which were both applied in clinical use. Complex 1 had intense antiproliferative activities against the human hepatoma cells(SMMC-7721)in vitro,which had the potential to develop as an anticancer drug in the future.As shown in Fig.9,the inhibition ratio of complex 1 were lower than the ratio of Na2(DCA)against human gastric cancer cells under the concentration of 300.00 μmol·L-1.In conclusion, the results showed complex 1 had strong antiproliferative effect against human hepatoma cells within the tested concentration range.It indicates that they had selectivity for the inhibition effect against cancer cells after forming complexes.
3 Conclusions
Two transition metal complexes[Ni(IM)(DCA) (H2O)2]·2H2O(1)and[Cd2(IM)4(DCA)2]·2H2O(2)were synthesized and characterized.Complex 1 was mononuclear molecule,and complex 2 was binuclear molecule.Central ion of each complex was sixcoordinated by nitrogen atoms from imidazoles and oxygen atoms from DCA.These complexes possess strong DNA and BSA binding abilities,and the binding intensity of complex 1 with DNA was stronger than complex 2.And the complex 1 had intense antiproliferative activities against the human hepatoma cells(SMMC-7721)in vitro,which gives it the potential to be developed as an anti-cancer drug in the future.
Acknow ledgements:We thank Institute of Zhejiang Academy of Medical Sciences for helping with anti-proliferative activity test.
[1]ZHOU Jian-Liang(周建良),CHUN Xiao-Gai(春晓改),ZHOU Lin-Jiao(周琳姣),et al.Chinese J.Inorg.Chem.(无机化学学报),2010,26(4):645-650
[2]Chen Z F,Liu Y C,Liu L M,et al.Dalton Trans.,2009,2: 262-272
[3]ZHANG Fan(张帆),LIN Qiu-Yue(林秋月),ZHENG Bo-Wen (郑博雯),et al.Chinese J.Inorg.Chem.(无机化学学报), 2012,28(11):2451-2457
[4]Lin Q Y,Wang Y Y,Feng Y L,et al.J.Coord.Chem.,2011, 64:920-930
[5]Zhang F,Zheng X L,Lin Q Y,et al.Inorg.Chim.Acta,2013, 394:85-91
[6]Timothy AH,ScottGS,ChristopherPG,etal.ChemMedChem, 2008,3(12):1878-1892
[7]Hart M E,Chamberlin A R,Walkom C,et al.Bioorg.Med. Chem.Lett.,2004,14:1969-1973
[8]To K K W,Ho Y P,Au-Yeung S C F.J.Chromatogr.A, 2002,947:319-326
[9]Semerci F,Yesilel Q Z,Sahin E.J.Inorg.Organomet.Polym. Mater.,2010,20:334-342
[10]YING Fu-Ling(尹富玲),SHEN Jia(申佳),ZOU Jia-Jia(邹佳嘉),et al.Acta Chim.Sinica(化学学报),2003,61:556-561
[11]Pang S K,Yu C W,Au-Yeung S C F,et al.B iochem.Biophys. Res.Commun.,2007,363:235-240
[12]Wang N,Lin Q Y,Wen Y H,et al.Inorg.Chim.Acta,2012, 384:345-351
[13]LI Shi-Kun(李士坤),LIN Qiu-Yue(林秋月),LÜ Tian-Xi(吕天喜),et al.Chinese J.Struct.Chem.(结构化学),2010,29: 1632-1637
[14]WANG Jun(汪俊),XU Qiong-Ming(许琼明),SUN Xiao-Fei (孙晓飞).Chin.J.Magn.Reson.(波谱学杂志),2009,26(1): 126-135
[15]Kumar C S A,Prasad S B B,Vinaya K,et al.Eur.J.Med. Chem.,2009,44:834-844
[16]Zheng X L,Sun H X,Liu X L,et al.Acta Pharmacol.Sin., 2004,25:1090-1095
[17]Sheldrick G M.SHELXS-97,Program for the Solution of Crystal Structures,University of Göttingen,Germany,1997.
[18]Sheldrick G M.SHELXL-97,Program for the Refinement of Crystal Structures,University of Göttingen,Germany,1997.
[19]Liu Z C,Wang B D,Yang Z Y,et al.Eur.J.Med.Chem., 2009,44:4477-4484
[20]WU Xiao-Yong(吴小勇),LIU Jian-Feng(刘建风),ZHAO Guo-Liang(赵国良).Chinese J.Inorg.Chem.(无机化学学报),2012,28(8):1661-1667
[21]Zhang F,Zhu W Z,Lin Q Y,et al.J.Rare Earths,2011,29 (4):297-302
[22]GUO Qiong(郭琼),LI Lian-Zhi(李连之),DONG Jian-Fang (董建方),et al.Acta Chim.Sinica(化学学报),2012,70:1617 -1624
[23]Lin Q Y,Wang Y Y,Feng Y L,et al.J.Coord.Chem.,2011, 64(5):920-930
[24]Lakowicz J R,Webber G.Biochemistry,1973,12(21):4161-4170
[25]Satish S B,Anupa A K,Hussain H,et al.Inorg.Chem., 2011,50:545-558
[26]Jose L G G,Javier H G,Aloma M R,et al.J.Inorg.Biochem., 2013,121:167-178
[27]Song W J,Cheng J P,Jiang W J,et al.Spectrochim.Acta A,2014,121:70-76
[28]Tan L F,Shen J L,Liu J,et al.Dalton Trans.,2012,41:4575 -4587
[29]Ashoka S,Seetharamappa J,Kandagal P B,et al.J.Lumin., 2006,121:179-186
[30]Wu X H,Liu J J,Wang Q,et al.Spectrochim.Acta A, 2011,79:1202-1209
[31]Guo Q,Li L Z,Dong J F,et al.Spectrochim.Acta A,2013, 106:155-162
[32]Wang Y J,Hu R D,Jiang D H,et al.J.Fluoresc.,2011,21: 813-823
[33]Patra A,Sarkar S,Mukherjee T,et al.Polyhedron,2011, 30:2783-2789
[34]Wang N,Lin Q Y,Feng J,et al.Inorg.Chim.Acta,2010, 363:3399-3406
Structures,Interaction w ith DNA and BSA and Antiproliferative Activities of Niand CdCom p lexes Based on Demethylcantharate and Im idazole
DU Fang-Yuan1,2LI Shi-Kun2LIN Qiu-Yue*,1,2WEI Qiong2TANG Ning-Ning2
(1Zhejiang Key Laboratory for Reactive Chemistry on Solid Surfaces,Zhejiang Normal University,Jinhua,Zhejiang 321004,China)
(2College of Chemical and Life Science,Zhejiang Normal University,Jinhua,Zhejiang 321004,China)
Two mixed-ligand complexes[Ni(IM)(DCA)(H2O)2]·2H2O(1)and[Cd2(IM)4(DCA)2]·2H2O(2)(DCA= demethylcantharate,7-oxabicyclo[2.2.1]heptane-2,3-dicarboxylate,C8H8O5;IM=imidazole,C3H4N2)were synthesized and characterized by elemental analysis,infrared spectra,thermogravimetric analysis,and X-ray diffraction. Complex 1 was a mononuclear molecule,with Niion six-coordinated by one nitrogen atom from imidazole, three oxygen atoms from DCA and two water molecules.The complex 1 crystallized in the monoclinic crystal system with P21/m.Complex 2 was a binuclear molecule,each Cdion was six-coordinated by two nitrogen atoms from two imidazole,one ether oxygen atom and three oxygen atoms of carboxyl groups from DCA.The complex 2 crystallized in the triclinic crystal system with P1 space group.The DNA binding properties of the complexes were investigated by electronic absorption spectra,fluorescence spectra and viscosity measurements. These two complexes could bind to DNA with moderate intensity via partial intercalation.The binding constants Kbwere 5.51×103(1)and 1.01×103L·mol-1(2)at 298 K,respectively.Meanwhile,the binding intensity of complexwith bovine serum albumin(BSA)is high,with binding constants KAequal to 1.91×105(1)and 6.17×105L·mol-1(2),respectively.Experimental results showed that complexes and BSA formed a 1∶1 compound with conformational changes in BSA.The antiproliferative activities of the complex(1)against human hepatoma cells (SMMC7721)lines and human breast cancer cells(MCF-7)lines were tested with MTT assay in vitro.The results showed that the inhibition effect had selectivity against cancer cells after forming complexes.The antiproliferative activities of the complex(IC50=(86.8±6.1)μmol·L-1)against the human hepatoma cells lines was more intense than Na2(DCA)(IC50=(152.8±15.6)μmol·L-1),which suggests potential application in anti-cancer drug development. CCDC:822330,1;822328,2.
imidazole;disodium demethylcantharate;nickel complex;cadmium complex;interaction with DNA and BSA; antiproliferative activity
O614.81+3;O614.24+2
A
1001-4861(2015)04-0813-11
10.11862/CJIC.2015.113
2014-10-20。收修改稿日期:2015-01-13。
国家自然科学基金(No.)资助项目。
*通讯联系人。E-mail:sky51@zjnu.cn

