1,3-二吗啉基丙烷与Hg和Zn的配合物:合成、结构与抗菌研究
Goudarziafshar Hamid Yousefi Somaieh Abbasityula YunesDušek Michal Eigner Václav Rezaeivala Majid Özbek Neslihan(Department of Chemistry,Faculty of Science,Sayyed Jamaleddinasadabadi University,Asadabad,Iran) (Department of Chemistry,Faculty of Science,Ilam University,P.O.Box 9 Ilam,Iran) (Department of Chemistry,Lorestan University,Khorramabad,Iran) (Institute of Physics AS CR,v.v.i.,Na Slovance ,8 Prague 8,Czech Republic) (Department of Chemical Engineering,Hamedan University of Technology,P.O.Box Hamedan,Iran) (Department of Primary Education,Faculty of Education,Ahi Evran University,000 Krsehir,Turkey)
Goudarziafshar Hamid*,1Yousefi Somaieh2Abbasityula Yunes3Dušek Michal4Eigner Václav4Rezaeivala Majid5Özbek Neslihan6
(1Department of Chemistry,Faculty of Science,Sayyed Jamaleddinasadabadi University,Asadabad,Iran) (2Department of Chemistry,Faculty of Science,Ilam University,P.O.Box 69315516 Ilam,Iran) (3Department of Chemistry,Lorestan University,Khorramabad,Iran) (4Institute of Physics AS CR,v.v.i.,Na Slovance 2,182 21 Prague 8,Czech Republic) (5Department of Chemical Engineering,Hamedan University of Technology,P.O.Box 65155 Hamedan,Iran) (6Department of Primary Education,Faculty of Education,Ahi Evran University,40100 Krsehir,Turkey)
以1,3-二吗啉基丙烷(DMP)为配体合成了2个配合物:Hg(DMP)Cl2(1)和Zn(DMP)Cl2(2),并对其结构进行了表征。单晶X-射线结构分析表明,2个配合物中的金属离子都与2个N原子和2个Cl原子配位,形成扭曲四面体配位结构。配体和配合物对3种革兰氏阳性菌 (S.aureus ATCC 25923,E.faecalis ATCC 23212和S.epidermidis ATCC 34384)和3种革兰氏阴性菌 (E.coli ATCC 25922,P.aeruginosa ATCC 27853和K.pneumonia ATCC 70063)都具有抗菌活性,且在一定条件下,Hg配合物的抗菌活性超过了作为标准的磺胺异恶唑药物。
晶体结构;1,3-吗啉基丙烷;抗菌活性;Hg配合物;Zn配合物
Morpholine is a strong alkali,which can be considered as a kind of secondary aliphatic amine. However,the incorporation of nitrogen into the sixmemberringexposestheloneelectronpairon nitrogen and makes the molecule a good nucleophile[1]. Morpholine and its derivatives are used extensively in many chemical reactions and its alkyl and aryl derivatives are very important in chemical and pharmaceutical industries[2-5].Heterocyclic ring systems with morpholine nucleus have aroused great interest in recent years due to their biological activities[6].Morpholine derivatives were reported to possess antimicrobial activity[7-9]and inflammatory activities in albino rats[10]. Morpholinederivativesexhibitanthelminticand insecticidal activity[11].N-substituted morpholines are used in the treatment of inflammatory diseases,asthma and migraine[12].N-substituted morpholines are also reported to show activity like antiemetics,platelet aggregation inhibitors and bronchodialators[13].Studies on the energetic of N-substituted morpholine derivatives are highly important in order to understand the biological activities and designing new drugs.
In continuation of our recent work on synthesis of the morpholine complexes[14],we report in this paper the synthesis and crystal structures of Hgand Zncomplexes of 1,3-dimorpholinopropane(DMP)ligand. Also,the antibacterial activity of the 1,3-dimorpholinopropane ligand and its metal complexes is reported against three Gram-positive species(S.epidermidis,S. aureus,E.faecalis)and three Gram-negative species (E.coli,P.aeruginosa,K.pneumonia)of bacterial strains by the disc diffusion and micro dilution methods.
1 Experimental
1.1Chemicals
All chemicals and salts of transition metals were of analytical grade(Sigma,Aldrich)and used as supplied.Solvents were distilled before use.
1.2Physical measurements
IR spectra of the ligand and complexes over the region of 400~4 000 cm-1were recorded using a Perkin Elmer spectrum(Version 10.01.00)FT-IR spectrometer with solid KBr pellets.1H NMR and13C NMR spectra were recorded on a BRUKER AC 400 MHz spectrometer using TMS as an internal standard in DMSO-d6solvent.Mass spectral data were obtained on Agilent Technology(HP).Elemental analyses were performed on a Vario ELⅢCHNS system.
1.3Synthesis
1.3.1Synthesis of 1,3-dimorpholinopropane(DMP)
The ligand was prepared according to the literature method[15].1,3-dibromopropane(4.69 g,25 mmol)in absolute ethanol(40 mL)was added drop wise to the mixture of potassium carbonate(6.91 g,50 mmol)and morpholine(4.35 g,50 mmol)in absolute ethanol(30 mL).The contents were refluxed for about 48 h at 80℃.The solution was then reduced to 20 mL by rotary evaporationandtheproductwasextractedwith chloroform.The product formed as a yellow Liquid, Yield:0.092 g,(43%).1H NMR(CDCl3,400 MHz):δ 1.3~1.7(m,-CH2-,2H);2.1~2.4(d,N-CH2,12H);3.47~3.61(d,O-CH2,8H);13C NMR(CDCl3,400 MHz):δ 65.4(d),55.4(c),52.2(b),22.1(a);IR(KBr,cm-1): 2 954 and 2 853(C-H);1 457(-CH2-);1 120(C-N);1 071 and 916(C-O).
1.3.2Syntheses of the complexes
Both complexes were synthesized by the branch tubemethod[16].1,3-dimorpholinopropane(DMP)(1 mmol,0.214 g)was placed in one arm of a branched tube and metal salts(1 mmol)in the other.Ethanol was then carefully added to fill both arms,the tube sealed and the ligand containing arm immersed in an oil bath at 60℃,while the other was left at ambient temperature.After one week,the colorless crystals were collected in the cooler arm.They were filtered off,washed with ether,and dried in air.For complexHg(DMP)Cl2(1):Yield:0.340 g(70%).Anal.Calcd. for C11H22Cl2N2O2Hg:C 27.2,H 4.5,N 5.8;Found:C 27.3,H 4.5,N 5.8.1H NMR(400 MHz,DMSO-d6):δ 1.81~1.82(m,-CH2-,2H);2.48~2.70(d,N-CH2,12H); 3.64~3.75(d,O-CH2,8H);13C NMR(400 MHz,DMSO -d6)δ 66.9(d);59.4(c);55.4(b);22.8(a);IR(KBr, cm-1):2 975 and 2 851(C-H);1 449(-CH2-);1 112(CN);1 078 and 910(C-O).For complex Zn(DMP)Cl2(2): Yield:0.228 g(65%).Anal.Calcd.for C11H22Cl2N2O2Zn: C 37.7,H 6.3,N 8.0;Found:C 37.6,H 6.2,N 7.9.1H NMR(400 MHz,DMSO-d6):δ 1.55~1.57(m,-CH2-, 2H);2.27~2.48(d,N-CH2,12H);3.45~3.54(t,O-CH2, 8H).13C NMR(400 MHz,DMSO-d6):δ 67.6(d);58.2 (c);55.1(b);24.4(a);IR(KBr,cm-1):2 969and 2 862(CH);1 464(-CH2-);1115(C-N);1 035 and 913(C-O).
1.4Single crystal X-ray structure determination
Single crystal X-ray diffraction intensity data of both complexes was performed at 120 K using Gemini four-circle diffractometer from Agilent Technologies, equipped with an area detector Atlas.Complex 1 was measured with Cu Kα radiation(λ=0.154 184 nm) monochromated and collimated with mirrors of Cu-Ultra collimator.For more absorbing complex 2 we used Mo Kα radiation(λ=0.071 07 nm)monochromated by graphite and collimated by Mo-Enhance collimator. In both cases,the source was a classical sealed X-ray tube.The structure was solved by charge flipping using SUPERFLIP program[17]and refined by fullmatrix least-square technique on F2,with JANA2006[18]program.Anisotropic displacement parameters were applied for all non-hydrogen atoms.The hydrogen atoms were discernible in difference Fourier maps,but according to common practice,H atoms bonded to C were kept in ideal positions with C-H=0.096 nm;the Uiso(H)was set to 1.2Ueq(C).The molecular structure plotswere prepared using the ORTEPⅢ[19]and Mercury[20].Crystallographic data and refinement parameters are summarized in Table1,and selected bond lengths and angles for complexes 1 and 2 are given in Table2.Selected non-classical hydrogen bonds are presented in Table3.
CCDC:957542,1;957846,2.

Table1 Crystal data and structure refinement of complexes 1 and 2

Continued Table1

Table2 Selected bond lengths(nm)and angles(°)for complexes 1 and 2

Table3 Intermolecular non-classical hydrogen bond in complexes 1 and 2
1.5Antimicrobial activity
The cultures of S.epidermidis ATCC 34384,S. aureus ATCC 25923,E.faecalis ATCC 29212,E.coli ATCC 25922,P.aeruginosa ATCC 27853,and K. pneumoniaATCC70063wereobtainedfromDepartment ofMedicalMicrobiology,HacettepeUniversity.Bacterial strains were cultured overnight at 37℃ in Nutrient Broth.During the survey,these stock cultures were stored in the dark at 4℃.
1.5.1Disc diffusion method
The synthesized compounds and complexes were dissolved in dimethylsulfoxide(20%DMSO)to a final concentration of 10 mg·mL-1and sterilized by filtration by 0.45 μm millipore filters.Antimicrobial tests were then carried out by the disc diffusion method using 100 μL of suspension containing 108CFU·mL-1bacteria,which was spread on a nutrient agar(NA) medium.The discs(6 mm in diameter)were impregnated with 25 μL of each compound(200 μg per disc)at the concentration of 8 mg·mL-1and placed on the inoculated agar.DMSO impregnated discs were used as negative control.Sulfioxazole(300 μg per disk)were used as positive reference standards to determine the sensitivity of one strain/isolate in each microbial species tested.The inoculated plates were incubated at 37℃for 24 h for bacterial strains isolates.Antimicrobial activity in the disc diffusion assaywasevaluatedbymeasuringthezoneof inhibition against the test organisms.Each assay in this experiment was repeated twice[21-22].
1.5.2Micro dilution assays
Theminimalinhibitionconcentration(MIC) values,except one,were also studied for the microorganisms sensitive to at least one of the four compounds determined in the disc diffusion assay.The inocula of microorganismswerepreparedfrom12hbroth culturesandsuspensionswereadjustedto0.5 McFarland standard turbidity.The test compounds dissolved in dimethylsulfoxide(DMSO)were first diluted to the highest concentration(2 mg·mL-1)to be tested,and then serial,two-fold dilutions were made in a concentration range from 0.015 625 to 2 mg·mL-1in 10 mL sterile test tubes containing nutrient broth. The MIC values of each compound against bacterial strainsweredeterminedbasedonamicro-well dilution method[23].The 96-well plates were prepared by dispensing 95 μL of nutrient broth and 5 μL of the inoculums into each well.100 μL from each of the test compounds initially prepared at the concentration of 2 mg·mL-1was added into the first wells.Then, 100 μL from each of their serial dilutions was transferred into eight consecutive wells.The last well containing195μLofnutrientbrothwithout compound,and 5 μL of the inoculums on each strip, was used as negative control.The final volume in each well was 200 μL.The contents of the wells were mixed and the micro plates were incubated at 37℃for 24 h.All compounds tested in this study were screened twice against each microorganism.The MIC wasdefinedasthelowestconcentrationofthe compounds to inhibit the growth of microorganisms[24].
2 Results and discussion
2.1Infrared and1H NMR spectra
In DMP,the infrared bands observed at 2 954, 2 853,1 457,1 071 and 916 cm-1were assigned to νC-H,νCH2
and νC-O-Cgroup respectively.A band due to νC-N-Cstretching vibration of the morpholine ring appeared at 1120 cm-1in the free ligand.This band was shifted to 1 112~1 115 cm-1in the metal complexes, suggesting the involvement of the nitrogen atom from the morpholine ring to the central metal ion.
Thus the infrared spectra strongly supported that the ligand coordinates through two nitrogen atoms while the two ether linkages are not involved in the complexation.
Evidence for the bonding mode of the 1,3-dimorpholinopropane was also provided by the1H NMR spectra of the free ligand and its complexes. The1HNMRspectrumofliganddisplayedthe following signals:-N-CH2protons at 2.1~2.4(d,12H), -O-CH2protons as doublet at 3.47~3.61(d,8H),-CH2-protons at 1.3~1.7(m,2H).The signals attributed to morpholineN-CH2protonsshifteddownfieldand appeared at 2.48~2.70 and 2.27~2.48 in the HgandZncomplexesrespectively.Whilesignals attributed to O-CH2groups slightly shifted rather than N-CH2protons(3.64~3.75 and 3.45~3.54 for Hgand Zncomplexes respectively).
2.2Crystal structure description of Hg(DMP)Cl2(1)
SinglecrystalX-rayanalysisrevealsthatcompound 1 crystallizes in monoclinic space group P21/c with one mercurycation,two chloride anions and one 1,3-dimorpholinopropane(DMP)ligand in the asymmetric unit.As shown in Fig.1,mercury is coordinated by two chlorines and two nitrogen atoms of the DMP ligand(HgN2Cl2)with distorted tetrahedral environment exemplified by its τ4parameter[25](τ4= [360-(a+b)]/141=0.83,where a and b are the two largest bond angles around the metal center).For ideal square and tetrahedral geometries the value of τ4would be zero and one,respectively.There are two different Hg-N bond lengths in the molecule;the shorter bond 0.235 7(3)nm is found for Hg(1)-N(10) while the longer one 0.239 4(3)nm is found for Hg(1)-N(6).The two Hg-Cl bond lengths are also different, 0.246 96(11)nm for Hg(1)-Cl(1)and 0.239 09(10)nm forHg(1)-Cl(2)(Table2).While there is no intramolecular interaction in crystal packing,its molecules areeventuallylinkedtogethertoformthreedimensional network via several inter-molecular nonclassical C-H…X(X=O,Cl)hydrogen bonds(Table3, Fig.2).In the complex 1,the morpholine rings adopt almost undistorted chair conformation with the torsion angles 57.30(43)°,60.16(42)°for N(10)O(13)C(11~15) and 58.64(50)°,58.66(46)°for N(6)O(3)C(1~5).
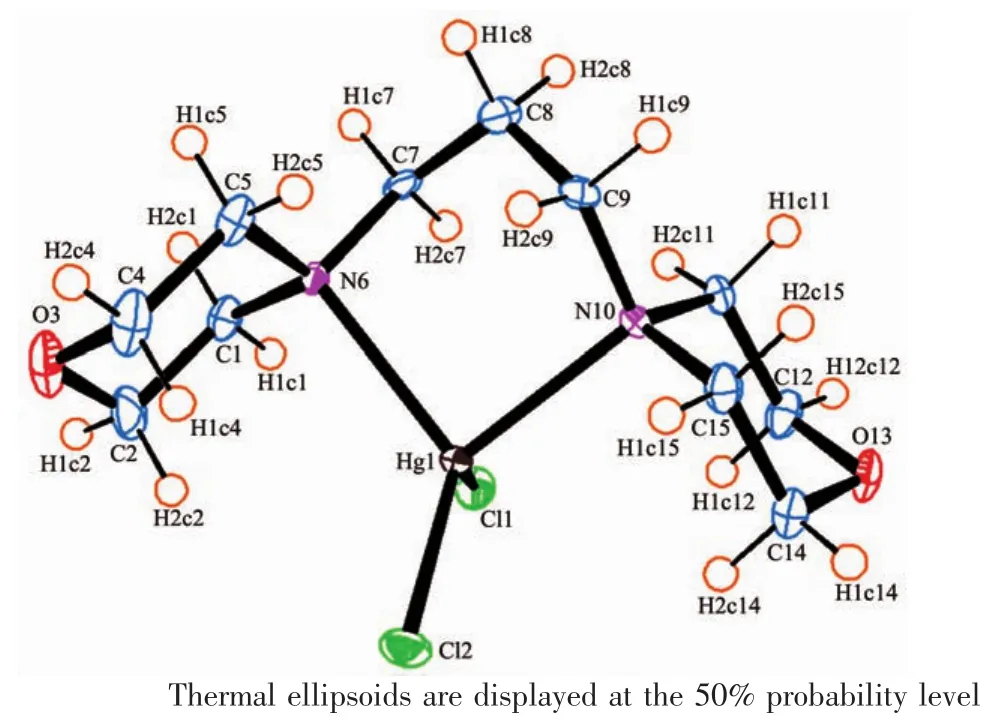
Fig.1 ORTEP diagram of 1 with atomic numbering scheme

Fig.2 Molecular packing of 1 viewed along a axis showing the molecules are linked into a 3D network
2.3Crystal structure description of[Zn(DMP)Cl2] (2)
Thetricliniccentrosymmetricstructureof2 containstwoindependentmoleculeswithslightly different geometry(Fig.3).The zinccenter is fourcoordinated by two chloride ions and two nitrogen atoms of the DMP ligand(ZnN2Cl2)in a distorted tetrahedral geometry(τ4=0.92 for Zn1a and 0.93 for Zn1b).

Fig.3 ORTEP drawing showing two independent molecules of 2
The endocyclic N6a-Zn1a-N10a angle(100.10(5)°) is narrower than the ideal tetrahedral angle of 109.5°, whereastheoppositeCl1a-Zn1a-Cl2aangle (115.129(18)°)is wider than the ideal tetrahedral angle (Table2).As expected,the morpholine ring adopts a chair conformation with torsion angles(56.42(18)°, 61.13(17)°),(57.81(19)°,61.21(18)°),(57.00(18)°, 60.57(18)°)and(57.56(19)°,60.69(19)°)for N(10a) O(13a)C(11~15a),N(6a)O(3a)C(1~5a),N(10b)O(13b) C(11~15b)and N(6b)O(3b)C(1~5b),respectively.Inthe crystal structure of 2,the molecules are linked into a three-dimensional network by the non-classical intermolecular C-H…O and C-H…Cl hydrogen bonds(Fig.4,Table3).

Fig.4 Molecular packing of 2,viewed along a axis, showing the molecules are linked into a 3D network
2.4Antibacterial activity results
The test compounds were screened in vitro for their antibacterial activity against three Gram-positive species(S.epidermidis,S.aureus,E.faecalis)and three Gram-negative species(E.coli,P.aeruginosa,K. pneumonia)of bacterial strains by the disc diffusion and micro dilutionmethods.Resulting antibacterial activities obtained by both methods are given in Table4 and Table 5.Also,the results were compared with those of the standard drug sulfioxazole are given in Fig.5 and 6.
The disc diffusion assay results evidently show (Table4)that Hg complex has strong inhibition effect against most of test bacteria whereas 1,3-dimorpholinopropaneligandandZncomplexhaveweaker activity.

Table4 Measured inhibition zone diameter(mm)of the complexes(200 μg per disk)and antibiotics by disc diffusion methodamm
According to the MICs results shown in Table 5, the compounds possess a broad spectrum of activitiesagainst the tested bacteria at the concentrations of 31.25~250 μg·mL-1.All compounds were active against P. aeruginosa ATCC 27853 at a concentration of 62.5~250 μg·mL-1whereas sulfisoxazole,the drug used as a standard,was found to be less active against the bacteria.HgcomplexshowedactivityagainstS. epidermidis ATCC 34384 at a concentration of 31.25~62.5 μg·mL-1and also in this case the activity of sulfisoxazole was lower.

Table 5 MICs of antibacterial activity of all complexes μg·mL-1

Fig.5 Comparison of antibacterial activity of ligand and its complexes with antibiotics
Inhibitionrateofthreecompoundswere exhibited in Fig.6.As shown in Fig.6,all compounds show excellent activity or good activity against P. aeruginosa.Hg complex also exhibits good activity against E.faecalis,whereas rest of the compounds only show significant activity.
3 Conclusions
Acknowledgments:Financial support for this work by the IlamUniversity,Ilam,Iranisgratefullyacknowledged. Crystallography part was supported by the project(Grant No.14-03276S)of the Czech Science Foundation.
[1]Jiang H,Zhang S,Xu Y.J.Mol.Struct.,2009,919:21-25
[2]Assaf G,Cansell G,Critcher D,et al.Tetrahedron Lett., 2010,52:5048-5051
[3]Madrakian T,Mohammadnejad M,Hojati F.J.Mol.Struct., 2010,968:1-5
[4]Shah N,Ul-Islam M,Khattak W A,et al.Carbohydr.Polym., 2013,98:1585-1598
[5]Jagtap R S,Joshi N N.Tetrahedron:Asymmetry,2011,22: 1861-1864
[6]Gnana Ruba Priya M,Panneerselvam P,Karikalan M.Int. J.Pharm.Biosci.,2011,2(1):267-272
[7]Basavaraja H S,Jayadevaiah K V,Mumtaz M H,et al.J. Pharm.Sci.Res.,2010,2:5
[8]Arjunan V,Rani T,Santhanalakshmi Mohan K S.Spectrochim.Acta A,2011,79(5):1386-1394
[9]Raparti V,Chitre T,Bothra K,et al.Eur.J.Med.Chem., 2009,44:3954-3960
[10]Xavier R J,Raj S A.Spectrochim.Acta A,2013,101:148-155
[11]Arjunan V,Rani T,Santhanalakshmi K,et al.Spectrochim. Acta A,2011,79:1395-1401
[12]Dugave C,Demange L.Chem.Rev.,2003,103:2475-2532
[13]Arrieta A,Oteagui D,Zubia A,et al.J.Org.Chem.,2007, 72:4313
[14]Goudarziafshar H,Rezaeivala M,Khosravi F,et al.J.Iran Chem.Soc.,2015,12:113-119
[15]UI-Haque M,Hussain M S,Ahmed J.J.Crystallogr.Spectrosc. Res.,1992,22:37-41
[16]Harrowfield J M,Miyamae H,Skelton B W,et al.Aust.J. Chem.,1996,49:1165-1169
[17]Palatinus L,Chapuis G.J.Appl.Crystallogr.,2007,40:786-790
[18]Petricek V,Dusek M,Palatinus L.JANA 2006,Structure Determination Software Programs,Institute of Physics,Praha, 2006.
[19]Farrugia L J.J.Appl.Crystallogr.,1997,30:565
[20]Macrae C F,Bruno I J,Chisholm J A,et al.J.Appl.Cryst., 2008,41:466
[21]Bauer A W,Kirby W M,Sherris J C,et al.Am.J.Clin. Pathol.,1966,45:493-496
[22]Gündüzalp A B,Özbay H.Russ.J.Inorg.Chem.,2012,57: 257-260
[23]Özbek N,Alyar S,Alyar H,et al.Spectrochim.Acta A, 2013,108:123-132
[24]Zhang F F,Gan L L,Zhou C H.Bioorg.Med.Chem.Lett., 2010,20:1881-1884
[25]Yang L,Powell D R,Houser R P.Dalton Trans.,2007:955-964
[26]Sultana N,Naz A,Arayne M S,et al.J.Mol.Struct.,2010, 969:17-24
Complexation of 1,3-Dimorpholinopropane with Hgand ZnSalts: Syntheses,Crystal Structures and Antibacterial Studies
Goudarziafshar Hamid*,1Yousefi Somaieh2Abbasityula Yunes3Dušek Michal4Eigner Václav4Rezaeivala Majid5Özbek Neslihan6(1Department of Chemistry,Faculty of Science,Sayyed Jamaleddinasadabadi University,Asadabad,Iran) (2Department of Chemistry,Faculty of Science,Ilam University,P.O.Box 69315516 Ilam,Iran) (3Department of Chemistry,Lorestan University,Khorramabad,Iran) (4Institute of Physics AS CR,v.v.i.,Na Slovance 2,182 21 Prague 8,Czech Republic) (5Department of Chemical Engineering,Hamedan University of Technology,P.O.Box 65155 Hamedan,Iran) (6Department of Primary Education,Faculty of Education,Ahi Evran University,40100 Krsehir,Turkey)
Two Hg(1)and Zn(2)complexes with general formulation M(DMP)Cl2where DMP=1,3-dimorpholinopropane were synthesized and structurally characterized by physicochemical methods and single crystal X-ray diffraction.An X-ray structural analysis shows that in both complexes Hg and Zn are coordinated by two nitrogen and two chlorine atoms in a form of distorted tetrahedron.The ligand and related complexes have antibacterial activity against three Gram-positive bacteria(S.epidermidis ATCC 25923,E.faecalis ATCC 23212 and S.epidermidis ATCC 34384),and also against the three Gram-negative bacteria(E.coli ATCC 25922,P.aeruginosaATCC 27853 and K.pneumonia ATCC 70063).The results revealed that in some cases the antibacterial activity of Hgcomplex exceeded the one of sulfisoxazole used as a standard.CCDC:957542,1;957846,2.
crystal structure;1,3-dimorpholinopropane;antibacterial activity;Hgcomplex;Zncomplex
O614.24+1;O614.24+3
A
1001-4861(2015)06-1076-09
10.11862/CJIC.2015.164
2015-01-26。收修改稿日期:2015-03-30。
Crystallography part was supported by the project(No.14-03276S)of the Czech Science Foundation.
*通讯联系人。E-mail:hamid-gafshar@yahoo.com