3株中国陕西盐湖耐金属离子嗜盐芽胞杆菌的分离鉴定
陈 锐,李 玥,孙晓宇,路鹏鹏,张 莉,沈卫荣*
(1.陕西微生物研究所微生物资源中心,陕西西安710043;2.陕西省质量技术监督局,陕西西安710000)
Moderately halophilic bacteria were heterogeneous physiological group of extreme microorganisms.Different genera and species were growth in saline environments[1].This kind of strain can accumulate and adjust the internal concentration of inorganic ions such as K+and Cl-to value that counteract the external osmolality[2].The first Halobacillus were described by Spring[3].There are 21 species have been recognized.It is believed that these microorganisms accumulate salt in the cytosol to counterbalance the external salt concentration[4-5].In order to investigate and collect the halophilic bacteria in the Northwest of China,400 halophilic strains were isolated from the salt lake waters or saline deposits of the HuaMaChi salt lake,which is in the arid region of shaanxi,China.HuaMaChi Salt Lake locates in the Loess Plateau and the Inner Mongolia Erdos desert zone.Around by Wind Beach landforms and border with Mu Us Desert(37°41'N 107°31'E).The isolated 400 strains belong to different genera and species.A375,A381 and A389 were considered to be Halobacillus-like strains by a 16S rRNA gene sequence analysis.The aims of the present study were to examine these three halophilic bacterial strain A375,A381 and A389 using polyphasic approach:includingphenotypic properties,chemotaxonomic properties,phylogenetic analysis which based on 16S rDNA gene sequences and metaltolerance test.
1 Materials and Methods
1.1 Bacterial strain
Bacterial strains A375,A381 and A389 were isolated by the dilution-plating technique from salt lake waters and saline deposits.
1.2 Culture medium
Complex Gibbons medium used for the isolation and maintenance of bacterial strains contained 7.5 g casein peptone,10 g yeast extract,2 g KCl,20 g MgSO4·7H2O,3 g sodium citrate and 100 g NaCl,dissolved in 1 L of water.If required,media were solidified by the addition of agar(20 g/L).The pH value was adjusted to 7.4.Unless indicated,all tests were carried out in medium with 10%NaCl(w/v),at pH 7.4 and incubated at 30℃.
Medium A contain 2 g(NH4)2SO4,100 g NaCl,500 mg NaH2PO4·H2O,500 mg K2HPO4,200 mg MgSO4·7H2O,100 mg CaCl2·2H2O,0.26 mg FeCl2,dissolved in 1 L of water.
1.3 Cell morphology and staining
Bacterial cultures were grown on Gibbons medium plates for 16 hours and then examined the Cell morphology by light microscopy(Leica).A.Gramstaining was performed according to Dussault[6].Photomicrographs of spores were obtained from cultures grown on Gibbons medium for 48 hours.Spore-staining was performed with 7.6%Malachite green for 10 minutes and staining with 0.5%Safranine O for 30 seconds.Colony morphology was examined for 72 hours incubation on Gibbons medium.Motility was assessed by using tubes containing medium Gibbons with agar(0.5%,w/v).Phenotypic tests were performed according to the proposed minimal standards for the description of novel taxa within the order Halobacteriales[7].
1.4 Tolerance of NaCl,pH and temperatures
Tolerance of NaCl and growth at various temperatures and pH were tested in Gibbons medium.Tolerance of NaCl was adjusting NaCl concentration range from 0~30%.The pH range for growth was determined by adjusting the pH to 5.6~11.0 with 1 mol/L NaOH.Growth tests were performed at the optimal growth temperature(30℃)for 2 days.The temperature range for growth was determined by incubation in liquid medium at temperatures between 16 to 50℃.
1.5 Hydrolysis and chemotaxonomic test
Gelatin hydrolysis was determined as described by Oren A,et al.[8].Hydrolysis of casein,L-Tyrosine or Starch was tested in medium A.Activities of Urase and Lipase were tested in medium Gibbons.For tests of nitrate reduction,medium A plus 0.1%(w/v)KNO3was employed.
1.6 Acid production
Growth on single carbon sources and produce acid was tested on liquid media A by added 1%the tested carbon sources compound,used Bromothymol-Blue as indicator.
1.7 Mentaltalerance of strain
Nine mental salt was used in this research:AgNO3,HgSO4,Pb(NO3)2,ZnSO4,K2Cr2O7,(CH3COO)2Cd·3H2O,CuSO4,CoCl3,MnSO4.Metaltolerance of the strain to metal salt was tested by add 5 μmol/L,10 μmol/L,20 μmol/L,50 μmol/L,0.1 mmol/L,0.2 mmol/L,0.5 mmol/L,1 mmol/L,2 mmol/L,10 mmol/L,20 mmol/L metal salt into Gibbons medium respectively and 200 r/min Shake culture at 30℃ for 24 h.Test bacterial suspension turbidity atOD600.If suspension greatly doped could assessment the strain growth was be inhibit.
1.8 Phylogenetic analysis of strains
The 16S rRNA gene sequence was determined by direct sequencing of the PCR products.Genomic DNA extraction was carried out as described by kit protocol(TIANGEN biotech DP302).PCR using the universal amplification primers 27F:5'-AGAGTTTGATCCTGGCTCA-3'and1492R:5'-GGTTACCTTGTTACGACTT-3'.Purification of the PCR products were carried out as described by kit protocol(TIANGEN biotech DP209).Sequence analyze were carry out by Sunnybio Company.The almost-complete 16S rRNA gene sequences of strains A375,A381 and A389 were compared with sequences from the GenBank,EMBL,DDBJ and PDBdatabases using the MEGABLAST program(NCBI).Alignmentofsequenceswascarried outusing CLUSTAL W software[9].Evolutionary distance matrices for the neighbour-joining method were calculated with the algorithm of Kimura[10]and the stability of the relationships was assessed by 1 000 bootstrap analyses.
2 Results
2.1 Cell morphology,Gram-staining and spore staining
Cells of strains were Gram-positive,spore-forming.The short rhabditiform of the cells remained the same in all phases of growth.After incubation for 48 h,all strains formed endospores located at central position(Fig.1).All strains were motile.After incubation for 72 h,diameter of colonies were about 3~4,2~3,2~3 mm respectively.Colonies of all strains were rounded,smooth and raised.Color of strain Colonies A385 were light yellow,A381 was pale yellow,and A389 was orange(Fig.2).

Fig.1 Gram stain and spore stain(Bars,20 μm)
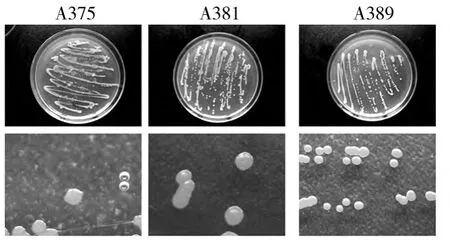
Fig.2 Strain A375,A381 and A389 colonial morphology
2.2 Tolerance of NaCl,pH,temperatures
A375 and A389 can grow in Gibbons liquid medium without Na+in 24 h.But Strain A381 required Na+for growth on Gibbons medium plate.Strain A375,A381 and A389 required Na+and Mg2+for growth,but was unable to grow when using K+instead of Na+or Br-instead of Cl-.Strain A375 grew in media containing 0~24%(w/v)NaCl,A381 at 5%~18%and A389 at 0~17%(w/v)NaCl.All strain optimal growth occurred with 10%(w/v)NaCl.
Strain A375,A381 and A389 growth maximum temperatures at 44,38,40℃ respectively,with optimum growth at 30℃.Strain A375,A381 and A389 grew in medium pH range 5.5~9.2.The optimum pH for growth was 8.0,7.5,and 7.5 respectively.
2.3 Acid produced,Hydrolysis and chemotaxonomic test
Acid was produced from D-fructose maltose,D-glucose,sucrose,D-trehalose and D-Mannitol,but not from D-mannose,sorbinse or D-xylose.
Casein was hydrolyzed by A375 and A381,but gelatin or L-tyrosine was not.Take discovered 18 kinds ofHabobacillussp.as reference,detailed results of morphological analyses and biochemical tests for strains A375,A381 and A389 are given in Table 1.
All strains are aerobic and Nitrate is not reduced to nitrite.Strain A375 was catalase,urase,oxidase,Lipase and VP test was positive but could not produced H2S.Strain A381 was catalase and urase test was positive.Lipase,H2S and VP test was negative.Strain A389 was catalase and lipase test posive.Urase,oxidase,H2S and VP test was negative.
2.4 Mentaltalerance
Add different kinds and certain concentrate of metal salt in Gibbons medium to testify strain A375,A381,A389 mental tolerance.Result show that add 0.01 mmol/L AgNO3,0.01 mmol/L HgSO4,0.02 mmol/L Pb(NO3)2,0.05 mmol/L(CH3COO)2Cd·3H2O,0.1 mmol/L ZnSO4,2 mmol/L K2Cr2O7,0.5 mmol/L CuSO4,0.05 mmol/L CoCl3or 10 mmol/L MnSO4in Gibbsons liquid medium will not influence A375 grow,butadd 0.02 mmol/L AgNO3,0.02 mmol/L HgSO4,0.05 mmol/L Pb(NO3)2,0.1 mmol/L(CH3COO)2Cd·3H2O,0.2 mmol/L ZnSO4,5 mmol/L K2Cr2O7,1 mmol/L CuSO4,0.1 mmol/L CoCl3or 20 mmol/L MnSO4in Gibbsons liquid medium,Growth of A375 was significantly inhibited.Results of strains A381 and A389 are given in Table 2.
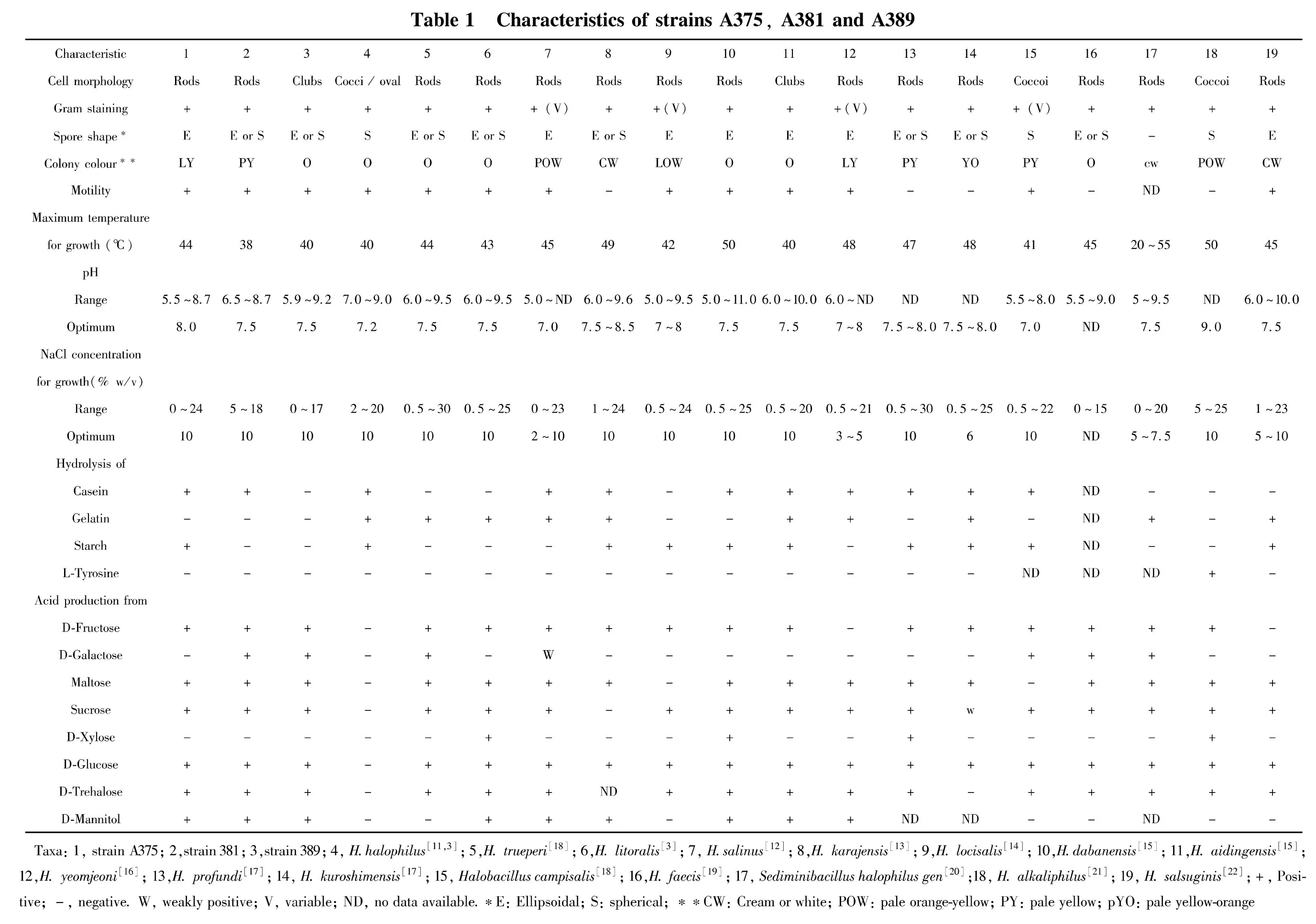
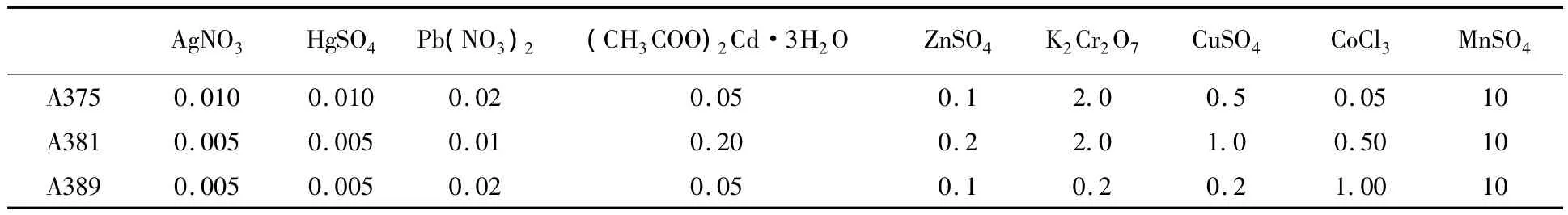
Table2 Metaltolerance of strains A375,A381 and A389(mmol/L)
2.5 Phylogenetic analysis of strains
The partial 16S rRNA gene sequence of strainsA375,A381 and A389 determined in this study comprised 1 436 nt.Comparative 16S rRNA gene sequence analyses showed that strain A375 was phylogenetically most closely affiliated to members of theHalobacillus kuroshimensis(Fig.1),A381 Affiliated toHalobacillus litoralis,A389 Affiliated toHalobacillus aidingensis.In the phylogenetic tree based on the neighbour-joining algorithm,strains A375,A381 and A389 fell within the radiation of the cluster comprisingHalobacillusspecies(Fig.3).The 16S rRNA gene sequence of strains A375,A381 and A389 showed similarity levels of 91.6%~98.7%with respect to sequences of the type strains of recognizedHalobacillusspecies(Fig.3).The almostcomplete 16S rRNA nucleotide sequences for strains A375,A381 and A389 were submitted to GenBank under the accession numbers JX415305,JX415307 and JX415308,respectively.

Fig.3 Neighbors-joining tree
3 Conclusion
A375,A381 and A389 strains are members of the genusHalobacillus,with a little bit difference from the typical strain on the basis of several phenotypic characteristics.However most of their characters are coincidence withHalobacillussp.A375 belongs toHalobacillus kuroshimensis,A381 is affiliated toHalobacillus litoralisand A389 is affiliated to
Halobacillus aidingensis.All these strains tolerance to the heavy mental salt.Further research need to be carried out aboutHalobacillussp.mentaltolerance mechanism.
[1]Ventosa A,Nieto J J,Oren A.Biology of moderately halophilic aerobic bacteria[J].Microbiol Mol Biol Rev,1998,62:504-544.
[2]Stephan H.Saum,Volker Müller.Salinity-Dependent Switching of Osmolyte Strategies in a Moderately Halophilic Bacterium:Glutamate Induces Proline Biosynthesis inHalobacillus halophilus[J].J Bacteriol,2007,189(19):6968-6975.
[3]Spring S,Ludwig W,Marquez,et al.Halobacillus gen.nov.,with descriptions ofHalobacillus litoralissp.nov.andHalobacillus trueperisp.nov.,and transfer ofSporosarcina halophilatoHalobacillus halophiluscomb.nov[J].Int J Syst Bacteriol,1996,46:492-496.
[4]Galinski E A,Trüper H G.Microbial behavior in salt-stressed ecosystems[J].FEMS Microbiol.Rev.,1994,15:95-108.
[5]Oren A,Bioenergetic aspects of halophilism[J].Microbiol.Mol.Biol.Rev,1999,63:334-348.
[6]Dussault H P.An improved technique for staining redHalophilic bacteria[J].J Bacteriol,1955,70:484-485.
[7]Oren A,Ventosa A,Grant W D.Proposal of minimal standards for the description of new taxa in the order Halobacteriales[J].Int J Syst Bacteriol,1997,47:233-238.
[8]Oren A,Elevi R,Watanabe S,et al.Halomicrobium mukohataeigen.nov.,comb.nov.,and emended description ofHalomicrobium mukohataei[J].Int J Syst Evol Microbiol,2002,52:1831-1835.
[9]Thompson J D,Higgins.CLUSTALW:improving the sensitivity of progressive multiple sequence alignment through sequence weighting,position-specific gap penalties and weight matrix choice[J].Nucleic Acids Res,1994,22:4673-4680.
[10]Kimura M.A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences[J].J Mol Evol,1980,16:111-120.
[11]Claus D,Fahmy F,Rolf HJ,et al.Sporosarcina halophilasp.nov.,an obligate,slightly halophilic bacterium from salt marsh soils[J].Syst Appl Microbiol,1983,4:496-506.
[12]Yoon Jung-Hoon,Kang Kook Hee,Park Yong-Ha.Halobacillus salinussp.nov.,isolated from a salt lake on the coast of the East Sea in Korea[J].Int J Syst Evol Microbiol,2003,3:687-693.
[13]Amoozegar M A,Malekzadeh F,Malik K A,et al.Halobacillus karajensissp.nov.,a novel moderate halophile[J].Int J Syst Evol Microbiol,2003,53:1059-1063.
[14]Yoon J-H,Kang K H,Oh T-K,et al.Halobacillus locisalissp.nov.,a halophilic bacterium isolated from a marine saltern of the Yellow Sea in Korea[J].Extremophiles,2004,8:23-28.
[15]Liu W Y,Zeng J,Wang L,et al.Halobacillus dabanensissp.nov.andHalobacillus aidingensissp.nov.,isolated from salt lakes in Xinjiang,China[J].Int J Syst Evol Microbiol,2005,55:1991-1996.
[16]Yoon Jung-Hoon,Kang So-Jung,Lee Choong-Hwan,et al.Halobacillus yeomjeonisp.nov.,isolated from a marine solar saltern in Korea[J].Int J Syst Evol Microbiol,2005,55:2413-2417.
[17]Hua Ngoc-Phuc,Kanekiyo Atsuko,Fujikura Katsunori,et al.Halobacillus profundisp.nov.andHalobacillus kuroshimensissp.nov.,moderately halophilic bacteria isolated from a deepsea methane cold seep[J].Int J Syst Evol Microbiol,2007,57:1243-1249.
[18]Yoon Jung-Hoon,Kang So-Jung,Jung Yong-Taek,et al.Halobacillus campisalissp. nov., containing mesodiaminopimelic acid in the cell-wall peptidoglycan,and emended description of the genusHalobacillus[J].Int J Syst Evol Microbiol,2007,57:2021-2025.
[19]An Sun-Young,Kanoh Kaneo,Kasaiet Hiroaki.Halobacillus faecissp.nov.,a spore-forming bacterium isolated from a mangrove area on Ishigaki Island,Japan[J].Int J Syst Evol Microbiol,2007,57:2476-2479.
[20]Carrasco I J,Mà rquez M C,Xue Y,et al.Sediminibacillus halophilusgen.nov.,sp.nov.,a moderately halophilic,Gram-positive bacterium from a hypersaline lake[J].Int J Syst Evol Microbiol,2008,58:1961-1967.
[21]Ida Romano,Ilaria Finore,Giancarlo Nicolaus,et al.Halobacillus alkaliphilussp.nov.,a halophilic bacterium isolated from a salt lake in Fuente de Piedra,southern Spain[J].Int J Syst Evol Microbiol,2008,58:886-890.
[22]Chen Yi-Guang,Zhang Yu-Qin,Liu Zhu-Xiang,et al.Halobacillus salsuginissp.nov.,a moderately halophilic bacterium from a subterranean brine[J].Int J Syst Evol Microbiol,2009,59:2505-2509.

