Association between endothelial nitric oxide synthase (ENOS) G894T polymorphism and high altitude (HA) adaptation: a meta-analysis
Hong-xiang LU, Yu-xiao WANG, Yu CHEN, Yong-jun LUO
1. Department of Military Medical Geography; College of High Altitude Military Medicine, the Third Military Medical University, Chongqing 400038, China; 2. Battalion 5 of Cadet Brigade, the Third Military Medical University, Chongqing 400038, China; 3. Key Laboratory of High Altitude Medicine (Ministry of Education); the Third Military Medical University, Chongqing 400038, China
Association between endothelial nitric oxide synthase (ENOS) G894T polymorphism and high altitude (HA) adaptation: a meta-analysis
Hong-xiang LU1,2,3, Yu-xiao WANG1,2,3, Yu CHEN3, Yong-jun LUO1,3
1. Department of Military Medical Geography; College of High Altitude Military Medicine, the Third Military Medical University, Chongqing 400038, China; 2. Battalion 5 of Cadet Brigade, the Third Military Medical University, Chongqing 400038, China; 3. Key Laboratory of High Altitude Medicine (Ministry of Education); the Third Military Medical University, Chongqing 400038, China
doi 10.13459/j.cnki.cjap.2015.06.006
Objective: Highland naves adapt well to the hypoxic environment at high altude (HA). Several genes have been reported to be linked to HA adaptaon. Previous studies showed that the endothelial nitric oxide synthase (ENOS) G894T polymorphism contributed to the physiology and pathophysiology of humans at HA by regulang the producon of NO. In this meta-analysis, we evaluate the associaon between the ENOS G894T polymorphism and HA adaptation through analyzing the published data. Methods: We searched all relevant literature about the ENOS G894T polymorphism and HA adaptaon in PubMed, Medline, and Embase before Step 2015. A random-ef f ects model was applied (Revman 5.0), and study quality was assessed in duplicate. Six studies with 634 HA nave cases and 621 low-altude controls were included in this meta-analysis. Results: From the results, we observed that the wild-type allele G was signif i cantly overrepresented in the HA groups (OR=1.85; 95% CI, 1.47-2.33;P<0.0001). In addion, the GG genotype was signif i cantly associated with HA adaptaon (OR=1.99; 95% CI, 1.54-2.57;P<0.0001). Conclusion: Our results showed that in 894 G allele carriers, the GG genotype might be a benef i cial factor for HA adaptaon through enhancing the level of NO. However, more studies were needed to conf i rm our fi ndings due to the limited sample size.
High altude (HA) adaptaon; endothelial nitric oxide synthase (ENOS); G894T polymorphism; meta-analysis
Introduction
Hypobaric hypoxia and cold were the typically environmental characteristics at high altitude (HA) [1-4]. Individual levels of physical and mental performance were reduced in such bad living environment[5]. While the original inhabitants living in plateau adapted to the extreme environment and performed well at high altitude for thousands of years, especially in three main HA areas: Tibetan Plateau (Tibetans, Ladakhi, and Sherpas), the Andes (Quechua and Ayamara), and Northern Africa
Received 2015-11-09; accepted 2015-11-20 (Ethiopians)[6]. Naturally, more attentions were paid to understand physiological reaction to hypoxia and whether HA natives had underwent natural selection[7]. During recent decades, a multitude of studies had been done to understand the human biology in HA natives. Several genes were recently reported to have relationships with HA adaptation, such as the mitochondrial DNA(mtDNA)[8, 9], Hypoxia-inducible factor-2alpha (HIF2A) [10], aldosterone synthase (CYP11B2)[11], the angiotensin-I converting enzyme (ACE)[12], endothelial nitric oxide synthase gene (ENOS)[13], hypoxia-inducible factor 1(HIF1A)[14].
Nitric oxide (NO), a gaseous signaling molecule involved in several physiological processes, was catalyzed by ENOS[15, 16]. A number of studies had investigated association of ENOS in HA adaptation, especially the polymorphisms of G894T (ENOS 894 G→T) [A nucleotide substitution leading to Asp re-place Glu at position 298 (Glu298Asp) in ENOS gene] [17]. Ahsan et al. reported an overrepresentation of wild-type alleles G of the ENOS gene were observed in Ladakh natives(over 3 600 m in India) and genotypes GG were benef i cial to HA adaptation[7]. In the meantime, Droma et al. demonstrated that Sherpa could adapt to HA better than non-Sherpa due to the higher frequencies of alleles G of the ENOS gene[18]. In addition, there were studies reported that NO levels were relevant to the G894T polymorphisms and the higher NO level benefited the highlanders’ HA adaptation[19]. Meanwhile, the meta-analysis made by Wang in 2013 showed the dominant ENOS G894T (Glu298Asp) SNP model (TT+GTvsGG) elevated high altitude pulmonary edema (HAPE) risk in Asians[20]. It was evident that polymorphisms of G894T had a great inf f uence on the regulation of HA physiology.
Higher NO levels in HA groups might be regulated by nature selection pressure, such as physical factors and hypoxia[21]. Consequently, the frequency of benef i cial alleles was represented dif f erently in the highlanders and the lowlanders[7].e wild-type alleles G of ENOS correlated with NO levels were signif i cantly higher in HA groups than in lowlanders, the more frequencies of genotype GG had an adaptive role at HA adaptation[7]. To address this issue, we performed this meta-analysis to assess the association between ENOS G894T polymorphism and HA adaptation.
Materials and Methods
Literature search
All literatures related to ENOS G894T polymorphism and HA adaptation were searched before Step 2015 using PubMed, Embase databases and Web of Science.e following keywords were used: ‘endothelial nitric oxide synthase polymorphism’’, “HA adaptation’’, “ENOS” and “adaptation”. Only articles limited to English were included, the search also excluded the abstracts and unpublished reports. If the same subjects were published in dif f erent publications, the study that was analyzed completely was extracted.is study accepted approval from the ethical committee of the Third Military Medical University in China.
Inclusion and exclusion criterca
Inclusion criteria were as following: (1) case-control study about the association between the ENOS G894T polymorphism and HA adaptation; (2) the outcome contained two comparison groups (adaptation groups vs. non-adaptation groups); (3) the papers provided data on the genotype number or frequency of ENOS; (4) HA cases were natives. Exclusion criteria were: (1) studies didn’t include genotype or frequency; (2) the outcome was not about relationship between ENOS and HA adaptation; (3)e ENOS G894T polymorphism in the controls did not accord with the Hardy-Weinberg equilibrium.
Data extraction
For each study, the first author, year of publication, altitude, study population and genotype number of cases and controls were recorded. All HA cases included in this meta-analysis were natives. Alleles G and T were computed from the similar genetic distribution. If one study provided multiple detailed data set about G894T polymorphism and HA adaptation, all the data was recorded. Finally, 21 unique references initially were extracted from the PubMed (11), Embase (6) and Web of Science (4). Of these, 14 publications were excluded according to criterca: provide no detailed data about ENOS genetype, without comparison on highlanders and lowlanders and multiple publications about the same data, 1 publication was excluded by researching the association between ENOS and high altitude diseases. Process of inclusion and exclusion was shown in Fig. 1.
Statistical analysis
In this meta-analysis, the association between the ENOS 894 G allele and adaptation of HA was evaluated by odds ratios (OR) and 95% conf i dence interval (CIs). We performed comparisons, for example, an allelic comparison (GvsT), and GG homozygous models (GGvsGT+TT, GGvsTT), GT heterozygous models (GTvsGG, GTvsTT), a TT homozygous model (GG+GTvsTT). Inconsistency in studies was calculated by the I-squared test. Heterogeneity was evaluated by the chi-square-based Q-test (It was significance whenP<0.05). When the heterogeneity was considered significant, we used a random effects model (DerSimonian-Laird method) to assess inter-study heterogeneity. The fixed effects model (Mantel-Haenszel) was calculated the pooled OR when there was no heterogeneity among the studies[22, 23]. We corrected for multiple hypothesis testing by the Bonferroni correction;P<0.008 has a significant difference. Statistical analyses were performed by Revman 5.0.
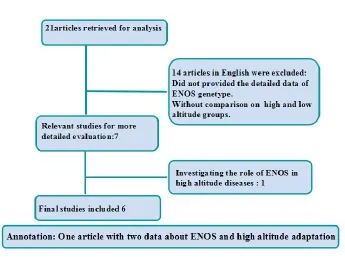
Fig. 1 Selection flow chart.
Results
Description of studies
A total of six studies with 634 cases and 621 controls were included. Especially, the study by Ahsan A (2005) provided two data sets considered to be independent studies. Baseline characteristics of the studies were presented in Table 1. Four studies focused on the natives in Asians (Sherpas, India), and two examined in America (Amerindian, Quechua). Adaptation ample sizes in the included papers ranged from 76 to 136, non-daptation ample sizes were 49 to 170 (Tab.1).
Meta-analysis results
In our study, the heterogeneity was observed among compare genotypes G versus T, GG versus GT+TT, GTvsGG (Tab.2). So the pooled odds ratio (OR) estimate for each study was calculated using the fi xed ef f ects model.e OR summary indicated signif i cant association between HA adaptation and carriers of the 894 G and T alleles (OR=1.85; 95% CI, 1.47-2.33;P<0.0001, Fig.2). Similarly, some association was observed for inheritance of genotypes (GGvsGT+TT, OR=1.99; 95% CI, 1.54-2.57;P<0.00001, Fig.3). Moreover, there was signif i cant correlation between having the genotype GTvsGG and HA adaptation (OR=0.52; 95% CI, 0.40-0.68;P<0.0001, Fig.4).
Due to this lack of heterogeneity compare genotypes GG versus TT, GG+ GTvsTT, GTvsTT, the pooled odds ratio (OR) estimate of each study was calculated by the fixed effects model(Tab.2). Genotypes showing signif i cant association were GG versus TT (OR=2.80; 95% CI, 1.26-6.26;P=0.01, Fig.5), and GG+ GTvsTT (OR=0.42; 95% CI, 0.19-0.94;P=0.03, Fig.6), Genotype (GTvsTT) showed no statistical signif i cance (OR=1.46; 95% CI, 0.65-3.03;P=0.36) between HA adaptation and the controls (Fig.7). According to the box plot of combined OR for fi gures 3-5, we could see that G allele was the protective allele (P50=1.46, Fig.8). However, just we can read from the table 1, the sample size of TT was far too small (Especially, in two of our studies there are⋆0⋆ TT homozygotes in the high altitude native category, and only one in the non-native category, the other studies are not much better), so these data of power in the TT allele category did not contribute to our analyses, those data should be excluded.
Discussion
In order to research the association of the ENOS G897T polymorphism with HA adaptation, we make this meta-analysis to provide a prospective study. A total of six published studies with 634 cases and 621 controls were included in this meta-analysis. We observed wild-type allele G were significantly more prevalent in the HA groups (OR=1.85; 95% CI, 1.47-2.33;P<0.00001). In addition, the GG genotype were greater in HA (OR=1.99; 95% CI, 1.54-2.57;P<0.00001). So we could make the conclusion that allele G and GG genotype may be benef i cial factors for HA adaptation.
It had been proposed that the high NO in the lung benef i ted human populations at HA[19].e hemoglobin oxygenation could be promoted by NO released in the lung, thus more oxygen was transported to the tissues[19]. Moreover, NO as a vasodilator played an important role in increasing systemic vasodilatation and pulmonary blood ff ow, thus reducing the scarcity of oxygen[19]. In addition, reports showed that inhalation of NO had been used to treat HA disorders[24]. All the observations supported the hypothesis that higher levels of NO would be benef icial to HA adaptation[7].
It had been reported that ENOS could be correlated to synthesis of NO via catalyzing the oxidation of the amino acid L-Arginine[15, 25]. Benef i cial alleles G enhanced the activity of ENOS enzyme and increased production of NO[25, 26]. Higher NO synthesis directly reduced endothelial dysfunction related to the development of HA disorders[27].us, alleles G of ENOS played key roles in catalytic enzymes increasing output of NO in the endothelial blood vessels[27]. Clearly, It could be supposed that those alleles G would be in favor of higher NO levels and genotype GG occurring significantly more frequently in natives may contribute to HA adaptation[7, 28].
In this meta-analysis, we proposed that G alleleand GG genotype of G894T benef i ted to HA adaptation. Meanwhile, available evidences indicated fewer wide-type alleles G damaged cardiovascular homeostasis that was important in human adaptation at HA[29]. Droma et al. (2002) also found that the rare wide alleles G in the ENOS gene were significantly more common in Japanese climbers who suffered HAPE, a life-threatening disease failing to adapt to HA[30]. Moreover, Ahsan et al. (2004) revealed that the Asp (T) alleles were over-represented in the HAPE groups in Indians[28].
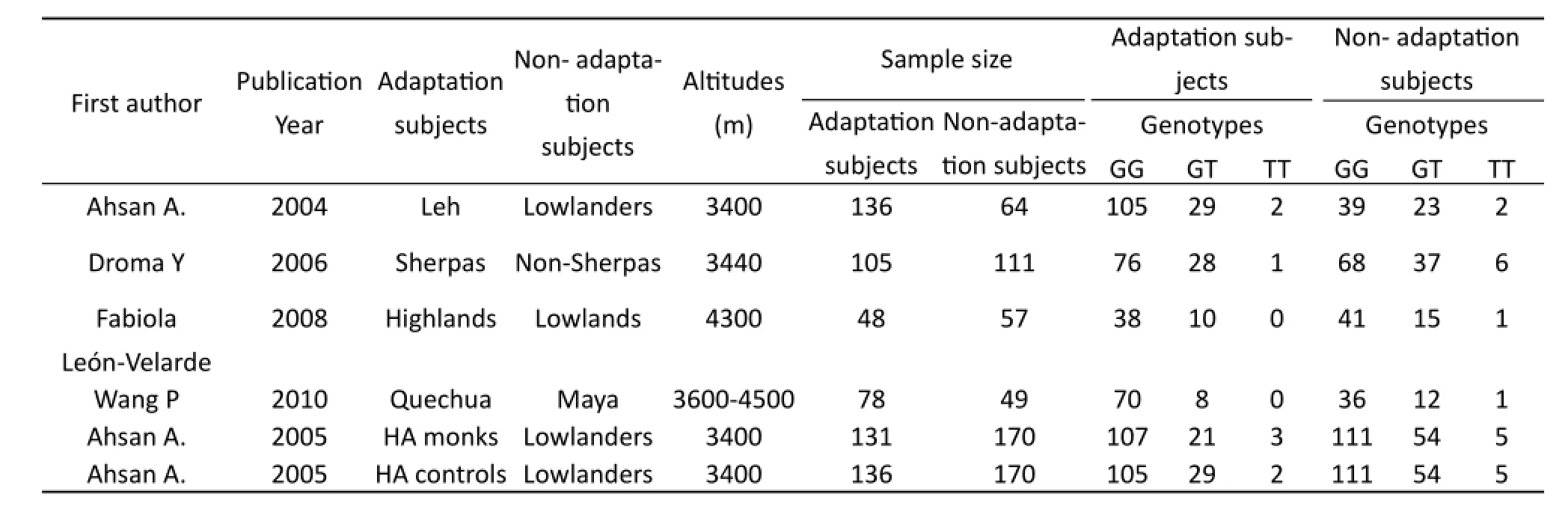
Tab. 1 Baseline characteristics of studies in the meta-analysis.
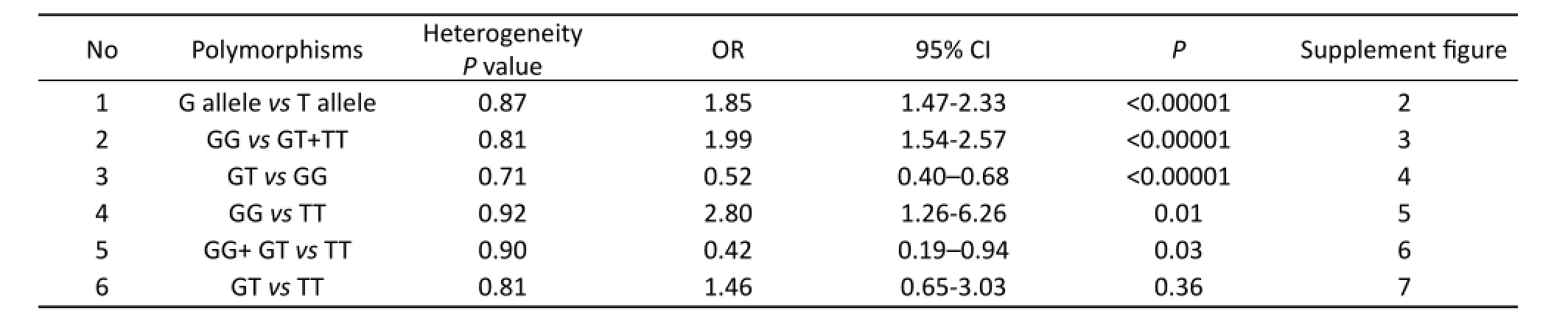
Tab. 2 Summary ORs and 95% confidence intervals of ‘G allele’ and ‘T allele’ in lowlanders and HA adaptation.

Fig. 2 Forest plot of ENOS G and T alleles with HA adaptation. The summary of genotype number or frequency (odds ratio) is displayed in the comparison of HA natives and lowlanders. The 95% confidence interval (95% CI) is shown on the extreme left and right.
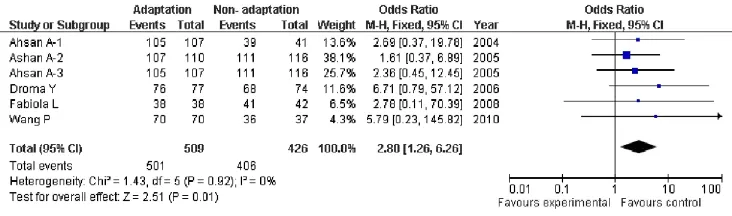
Fig. 3 Forest plot of ENOS GGvsGT +TT with HA adaptation. The summary of genotype number or frequency (odds ratio) is displayed in the comparison of HA natives and lowlanders. The 95% confidence interval (95% CI) is shown on the extreme left and right.

Fig. 4 Forest plot of ENOS GGvsGT with HA adaptation. The summary of genotype number or frequency (odds ratio) is displayed in the comparison of HA natives and lowlanders. The 95% confidence interval (95% CI) is shown on the extreme left and right.

Fig. 5 Forest plot of ENOS GGvsTT with HA adaptation. The summary of genotype number or frequency (odds ratio) is displayed in the comparison of HA natives and lowlanders. The 95% confidence interval (95% CI) is shown on the extreme left and right.
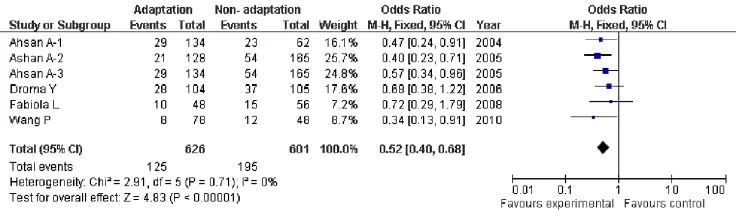
Fig. 6 Forest plot of ENOS GG+GTvsTT with HA adaptation. The summary of genotype number or frequency (odds ratio) is displayed in the comparison of HA natives and lowlanders. The 95% confidence interval (95% CI) is shown on the extreme left and right.
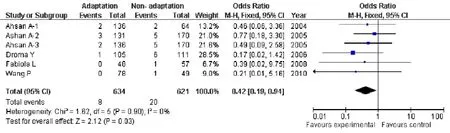
Fig. 7 Forest plot of ENOS GTvsTT with HA adaptation. The summary of genotype number or frequency (odds ratio) is displayed in the comparison of HA natives and lowlanders. The 95% confidence interval (95% CI) is shown on the extreme left and right.

Fig. 8 Box plot of combined odds ratios for figures 3-5.
Although this meta-analysis supported the hypothesis that alleles G of the ENOS gene benefited altitude tolerance, the following limitations should be considered. First, the natives included were located in different altitude and came from different race, thus might af f ect the outcomes of our meta-analysis, although a random-ef f ects model had been applied. Second, the level of NO in the lung wasn’t measured due to logistical dif ficulties in Quechua.ird, relative small sample size limited the statistical power,a great number of cases should be researched to explore the HA adaptation. Finally, other relevant genes involved in HA adaptation was not presented.
Conclusion
We analyzed the association of the 894G/T polymorphism with HA adaptation, and concluded that in 894 G allele carriers, GG might be a beneficial factor for HA adaptation. However, the results of meta-analysis were unavailable due to the limited data. More relevant studies should be performed to confi rm the association between894G/T polymorphism and HA adaptation in the future.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81372125).
Disclosures
1. Ward M P. Everest 1953, first ascent: a clinical record[J]. High Alt Med Biol, 2003, 4(1): 27-37.
2. Tang E, Chen Y, Luo Y. Dexamethasone for the prevention of acute mountain sickness: Systematic review and meta-analysis[J]. Int J Cardiol, 2014, 173(2): 133-138.
3. Luo Y, Yang X, Gao Y. Strategies for the prevention of acute mountain sickness and treatment for large groups making a rapid ascent in China[J]. Int J Cardiol, 2013, 169(2): 97-100.
4. Luo Y, Chen Y, Zhang Y,et al.e association of angiotensin-converting enzyme gene insertion/ deletion polymorphisms with acute mountain sickness susceptibility: a meta-analysis[J]. High Alt Med Biol, 2012, 13(4): 252-257.
5. Qadar Pasha MA, Khan AP, Kumar R,et al. Angiotensin converting enzyme insertion allele in relation to high altitude adaptation[J]. Ann Hum Genet, 2001, 65(Pt 6): 531-536.
6. Stobdan T, Karar J, Pasha MA. High altitude adaptation: genetic perspectives[J]. High Alt Med Biol, 2008, 9(2): 140-147.
7. Ahsan A, Norboo T, Baig MA,et al. Simultaneous selection of the wild-type genotypes of the G894T and 4B/ 4A polymorphisms of NOS3 associate with high-altitude adaptation[J]. Ann Hum Genet, 2005, 69(Pt 3): 260-267.
8. Luo Y, Yang X, Gao Y. Mitochondrial DNA response to high altitude: a new perspective on high-altitude adaptation[J]. Mitochondrial DNA, 2013, 24(4): 313-319.
9. Luo Y, Gao W, Liu F,et al. Mitochondrial nt3010G-nt3970C haplotype is implicated in high-altitude adaptation of Tibetans[J]. Mitochondrial DNA, 2011, 22(5-6): 181-190.
10. Yi X, Liang Y, Huerta-Sanchez E,et al. Sequencing of 50 human exomes reveals adaptation to high altitude[J]. Science, 2010, 329(5987): 75-78.
11. Rajput C, Arif E, Vibhuti A,et al. Predominance of interaction among wild-type alleles of CYP11B2 in Himalayan natives associates with high-altitude adaptation[J]. Biochem BiophysRes Commun, 2006, 348(2): 735-740.
12. Myerson S, Hemingway H, Budget R,et al. Human angiotensin I-converting enzyme gene and endurance performance[J]. J Appl Physiol, 1999, 87(4): 1313-1316.
13. Rajput C, Najib S, Norboo T,et al. Endothelin-1 gene variants and levels associate with adaptation to hypobaric hypoxia in high-altitude natives[J]. Biochem Biophys Res Commun, 2006, 341(4): 1218-1224.
14. Liu KX, Sun XC, Wang SW,et al. [Association of polymorphisms of 1772 (C-->T) and 1790 (G-->A) in HIF1A gene with hypoxia adaptation in high altitude in Sherpas][J]. Chin J Med Genet, 2007, 24(2): 230-232.
15. Wang P, Ha AY, Kidd KK,et al. A variant of the endothelial nitric oxide synthase gene (NOS3) associated with AMS susceptibility is less common in the Quechua, a high altitude Native population[J]. High Alt Med Biol, 2010, 11(1): 27-30.
16. Luo Y, Chen Y, Zhang Y,et al. Association of endothelial nitric oxide synthase (eNOS) G894T polymorphism with high altitude pulmonary edema susceptibility: a meta-analysis[J]. Wilderness Environ Med, 2012, 23(3): 270-274.
17. Marsden PA, Heng HH, Scherer SW,et al. Structure and chromosomal localization of the human constitutive endothelial nitric oxide synthase gene[J]. J Biol Chem, 1993, 268(23): 17478-17488.
18. Droma Y, Hanaoka M, Basnyat B,et al. Genetic contribution of the endothelial nitric oxide synthase gene to high altitude adaptation in sherpas[J]. High Alt Med Biol, 2006, 7(3): 209-220.
19. Beall CM, Laskowski D, Strohl KP,et al. Pulmonary nitric oxide in mountain dwellers[J]. Nature, 2001, 414(6862): 411-412.
20. Wang QQ, Yu L, Huang GR,et al. Polymorphisms of angiotensin converting enzyme and nitric oxide synthase 3 genes as risk factors of highaltitude pulmonary edema: a case-control study and meta-analysis[J]. Tohoku J Exp Med, 2013, 229(4): 255-266.
21. Chatterjee A, Black SM, Catravas JD. Endothelial nitric oxide (NO) and its pathophysiologic regulation[J]. Vascul Pharmacol, 2008, 49(4-6): 134-140.
22. Ma X, Chen C, Xiong H,et al. Transforming growth factorbeta1 L10P variant plays an active role on the breast cancer susceptibility in Caucasian: evidence from 10,392 cases and 11,697 controls[J]. Breast Cancer Res Treat, 2010, 124(2): 453-457.
23. Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease[J]. J Natl Cancer Inst, 1959, 22(4): 719-748.
24. Hackett PH, Roach RC. High-altitude illness[J]. N Engl J Med, 2001, 345(2): 107-114.
25. Garg UC, Hassid A. Nitric oxide-generating vasodilators and 8-bromo-cyclic guanosine monophosphate inhibit mitogenesis and proliferation of cultured rat vascular smooth muscle cells[J]. J Clin Invest, 1989, 83(5): 1774-1777.
26. Wang XL, Mahaney MC, Sim AS,et al. Genetic contribution of the endothelial constitutive nitric oxide synthase gene to plasma nitric oxide levels[J]. Arterioscler Thromb Vasc Biol, 1997, 17(11): 3147-3153.
27. Leon-Velarde F, Mejia O. Gene expression in chronic high altitude diseases[J]. High Alt Med Biol, 2008, 9(2): 130-139.
28. Ahsan A, Charu R, Pasha MA,et al. eNOS allelic variants at the same locus associate with HAPE and adaptation[J]. Thorax, 2004, 59(11): 1000-1002.
29. Groves BM, Droma T, Sutton JR,et al. Minimal hypoxic pulmonary hypertension in normal Tibetans at 3,658 m[J]. J Appl Physiol (1985), 1993, 74(1): 312-318.
30. Droma Y, Hanaoka M, Ota M,et al. Positive association of the endothelial nitric oxide synthase gene polymorphisms with high-altitude pulmonary edema[J]. Circulation, 2002, 106(7): 826-830.
Yong-jun LUO, College of High Altitude Military Medicine, the Third Military Medical University, Chongqing 400038, China; Tel: 86-23-68752396; Fax: 86-23-68752396; E-mail: luoyongjun2011@gmail.com
- 中国应用生理学杂志的其它文章
- A mini review: Tau transgenic mouse models and olfactory dysfunction in Alzheimer’s Disease
- Ethical inspection about laboratory animals
- Better parameters of ventilation-CO2output relationship predict death in CHF patients
- Flow cytometric analysis of circulating microvesicles derived from myocardial ischemic preconditioning and cardioprotection of ischemia/reperfusion injury in rats
- Synergisms of cardiovascular effects between iptakalim and amlodipine, hydrochlorothiazide or propranolol in anesthetized rats
- Effects of curcumin on sodium currents of dorsal root ganglion neurons in type 2 diabetic neuropathic pain rats

