Expression Analysis of the Insulin-Like Growth Factors I and II During Embryonic and Early Larval Development of Turbot (Scophthalmus maximus)
WEN Haishen, QI Qian, HU Jian, SI Yufeng, HE Feng, and LI Jifang
Fisheries College,Ocean University of China,Qingdao266003,P. R. China
Expression Analysis of the Insulin-Like Growth Factors I and II During Embryonic and Early Larval Development of Turbot (Scophthalmus maximus)
WEN Haishen*, QI Qian, HU Jian, SI Yufeng, HE Feng, and LI Jifang
Fisheries College,Ocean University of China,Qingdao266003,P. R. China
The insulin-like growth factors I and II (IGF-I and IGF-II) are important proteins involved in fish growth and development. Here, we report the isolation of IGF-II and expression analysis of IGFs in turbotScophthalmus maximus, aiming to clarify their function in embryonic and larval development of fish. The deduced IGF-II gene is 808 bp in full length, which encodes a protein of 219 amino acids and is 93% similar with that ofParalichthys olicaceusin amino acid sequence. The tissue abundance and the expression pattern of IGFs in a turbot at early development stages were investigatedviareverse transcription-polymer chain reaction. Result showed that the IGF-I and IGF-II genes were widely expressed in tissues ofS. maximus. IGF-I was detected in all tissues except intestines with the highest level in liver, while IGF-II transcript presented in all tissues except muscle. At the stages of embryonic and larval development, the mRNA levels of IGFs sharply increased from the stage of unfertilized egg to post larva, followed by a decrease with larval development. However, there was an increase in IGF-I at the embryonic stage and IGF-II at the gastrula stage, respectively. These results suggested that IGFs play important roles in cell growth and division of the turbot. Our study provides reference data for further investigation of growth regulation in turbot, which can guarantee better understanding of the physiological role that IGFs play in fish.
Scophthalmus maximus; insulin-like growth factor; cloning; expression analysis
1 Introduction
The insulin-like growth factors I and II (IGF-I and IGF-II) are two highly homologous mitogenic polypeptides which function ubiquitously and show diverse effects on the development, growth, and metabolism of animals (reviewed by Werner and LeRoith, 1996). The IGF system, including IGF receptor and its binding protein (IGFBP), plays important roles in regulating vertebrate growth (Reinecke and Collet, 1998). IGFs are single-chain polypeptides initially translated as prepropeptides with a leader signal peptide at N-terminal, which is followed by domains B, C, A, and D and domain E trailer at C-terminal (Tseet al., 2002).
In early researches, the IGF genes have been cloned in many vertebrates, and their expression modes have been studied. The evidenced is that teleost IGFs are initially produced in liver, and hepatic tissues synthesize IGFs as well. The IGFs act through local paracrine and autocrine pathways as sensitive regulators of testicular function (Reinecke and Collet, 1998; Gaoet al., 2012). IGF-I hasbeen shown to exhibit a wide range of biological actions including stimulation of cell division and differentiation as well as protection from protein degradation and apoptosis. In addition, IGF-I acts as a regulator of endocrine factors such as growth (Jones and Clemmons, 1995; Uptonet al., 1998). IGF-II is a potent mitogenic and survival factor involved in the regulation of growth, development, and reproduction in animals (Yuanet al., 2011).
The IGF-I cDNA sequence has been cloned in a number of teleosts including chinook salmon (Wallis and Devlin, 1993), coho salmon (Duguayet al., 1992), catfish (McRory and Sherwood, 1994), gilthead seabream (Duguayet al., 1996), goldfish (Kermounet al., 1998), and turbot (Duvalet al., 2002). IGF-II amino acid sequence was first reported in human in 1978 (Rinderknechtet al., 1978), and then found in many fish species. Though Duvalet al. (2002) have determined the partial sequence of IGF-II in turbot, it is meaningful to further study the full-length IGF-II in this species for better understanding its physiological function. Despite the knowledge that IGFs play crucial roles in the growth of mammals (De Pabloet al., 1993; Rosenet al., 1993; Scavoet al., 1991), information is lacking on the function of the IGF system in the early development of fish. Duguayet al. (1996) found that IGFs are expressed in gilthead seabream dur-ing larval development, whereas Greene and Chen (1997) suggested that IGFs are expressed in embryos of rainbow trout. Despite a study on the regulation between IGFs and growth hormone (GH) release in turbot (Duvalet al., 2002), the expression pattern of IGFs genes during the embryonic and larval developmental stages of this species is unknown.
The turbotScophthalmus maximusis an important commercial flatfish species inhabiting European waters. It was first introduced into China for farming in 1992, and as a result of successful artificial breeding, commercial culture of the species has spread rapidly along the coast of North China since 1999 (Lei and Zhan, 2001). Previously, we have cloned the IGFBP-1 and -2 genes and analyzed their expression at the stages of adult and early development (Huet al., 2012). In the present study, we cloned IGF-II gene from liver ofS. maximusand analyzed the expression of IGF-I and IGF-2 genes during the stages of embryogenesis and early larval development in order to understand how the growth of turbot is regulated. The results will contribute to understanding the physiological characteristics ofS. maximusand providing a reference data for rational breeding of this fish species.
2 Materials and Methods
2.1 Experimental Fish
Adult turbot (S.maximus) were obtained from coastal area of Shandong Province during a period from September 2009 to next May. The fish were anesthetized with MS-222 (Sigma, St. Louis, MO, USA) and sacrificed after decapitation. Tissues including ovary, intestine, muscle, gills, kidney, liver, stomach, heart, brain, spleen, and skin were collected from adult turbot and immediately frozen in liquid nitrogen, then stored at -80℃ till use. Additionally, unfertilized eggs (0-hour-post-fertilization, 0HPF); fertilized eggs in the gastrula (18HPF), embryonic (80HPF) and hatching (120HPF) stages; and the prophase of hatching (13-day-post-hatching, 13DPH) and post larva (25DPH) were obtained from a commercial hatchery and immediately frozen in liquid nitrogen, then stored at -80℃ till use.
2.2 Total RNA Extraction and Reverse Transcription
Total RNA was extracted using Trizol reagent (Invitrogen, USA). The intactness and purity of RNA extracts were determined with a UV spectroscopy at 260 and 280 nm. Total RNA (1 μg) was subjected to reverse transcription in a total volume of 10 μL with random primers and M-MLV reverse transcriptase (Takara, Japan).
2.3 Molecular Cloning and Sequence Characterization of IGF-II cDNA
The strategy for isolating IGF-II full length cDNA involved the generation of a partial cDNA from liver by reverse transcription-polymerase chain reaction (RT-PCR) with degenerate primers firstly, then the amplification of 5’ and 3’-cDNA ends by rapid amplification of cDNA ends (RACE), and finally the generation of a cDNA that encodes the complete coding region in a single set of RT-PCR. For IGF-II gene fragment amplification, the primers IGF2f and IGF2r (Table 1) were designed using the IGF-II amino acid sequences of tilapia (Oreochromis niloticus), gilthead seabream (Sparus aurata), rainbow trout (Oncorhynchus mykiss), and European seabass (Dicentrarchus labrax). The 50-µL PCR reaction contained 2 μL of cDNA from liver tissue following the manufacturer’s instructions (Takara, Japan). PCR amplification was performed using the following touchdown PCR cycling conditions: pre-denaturation at 94℃ for 5 min; 10 cycles of 94℃ for 30 s, 65℃ (decreasing by 1℃each cycle) for 30 s, and 72℃ for 30 s, followed by additional 25 cycles of 94℃ for 30 s, 55℃ for 30 s and 72℃for 30 s; and final extension at 72℃ for 10 min. According to the IGF-II fragment, PCR was performed with the 5’and 3’ RACE primers and nested-PCR primers (listed in Table 1) designed using Primmer5.0 and DNAMAN software; the 5’ and 3’ RACE reactions were obtainedwith a SMARTTMRACE cDNA Amplification Kit (Clontech, USA). Products of the degenerate PCR and RACE reactions were electrophoresed on 1.5% agarose gel. The DNA band of expected size was purified with a TIAN Gel Extraction Kit (TIANGEN, China). The purified DNA fragments were then cloned into PGM-T vector (TIANGEN, China), propagated inE. coliDH5, and sequenced using an ABI3730XL sequencer.

Table 1 Primers used forScophthalmus maximusIGF-II gene cloning and IGFs (I and II) gene expression analysis
2.4 Phylogenetic Analysis and Sequence Analysis
The presence and location of putative signal peptide cleavage sites and potential N-glycosylation sites in the amino acid sequences were predicted using the prediction servers of the Center for Biological Sequence Analysis (http://www.cbs.dtu.dk/services/). Amino acid sequences were retrieved from GenBank (Altschulet al., 1990), and the sequences of IGF-II were aligned with the homologs of other fish. Multiple protein sequence alignments were performed in Clustal X 1.81 (Thompsonet al., 1997). Phylogenetic analyses of full-length amino acid sequences were conducted using MEGA 2.0 (Tamuraet al., 2007). A rooted phylogenetic tree was constructed using the neighbor-joining algorithm (Saitou and Nei, 1987), and the data were re-sampledvia1000 bootstrapping replicates.
2.5 RT-PCR Analysis
The mRNA abundances of IGFs genes were detected by semi-quantitative RT-PCR assays. Tissue expression of IGFs genes was detected with a mature-stage sample. Total RNA extracts from the tissues, including ovary, intestine, muscle, gills, kidney, liver, stomach, heart, brain, spleen, and skin, were treated as described above. Temporal expression of IGFs genes was investigated in unfertilized and fertilized eggs at the gastrula (18HPF), embryonic (80HPF), and hatching (120HPF) stages, prophase of hatching (13DPH) and post larva (25DPH).
RT-PCR was carried out using a Takara TaqTMkit (Takara, Japan) according to the manufacturer’s instructions. The PCR program was set as follows: degeneration at 94℃ for 5 min, followed by 34 cycles of 94℃ for 30 s, 62℃ for 30 s (both IGFs), 72℃ for 30 s, and final extension at 72℃ for 10 min (primers listed in Table 1), using 18S rRNA as an internal standard. The PCR program of 18S rRNA was 94℃ for 5 min, followed by 25 cycles of 94℃ for 30 s, 61℃ for 30 s, and 72℃ for 30 s, and final extension at 72℃ for 10 min. Each reaction product was resolved on a 1.5% agarose gel containing ethidium bromide and visualized on a gel system (Tanon, China).
3 Results
3.1 Characteristics of IGF-II cDNA
The cloned IGF-II cDNA (GenBank Accession No.:JN032705) is 880 bp in full length, which contains a 165-bp 5’-untranslated terminal region (UTR), a 55-bp 3’-UTR, and a 657-bp open reading frame. It encodes a protein of 219 amino acids, and the amino acid contains a signal peptide and domains B, C, A, D, and E with 6 conserved cysteine residues in domain B and domain A (Fig.1).
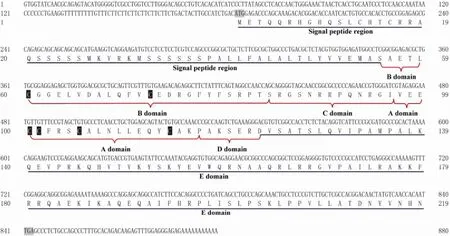
Fig.1 Full sequence ofScophthalmus maximusIGF-II. The domains B, C, A, D, and E are shown after the signal peptide region; 6 cysteine residues are indicated by black background and white letters.
3.2 Phylogenetic Relationship
Results of the sequence comparison analysis showed that the IGF-II gene of turbot is highly similar with those of redbanded seabream (Pagrus auriga), gilthead seabream (Sparus aurata), white seabream (Diplodus sargus), andorange-spotted grouper (Epinephelus coioides) (Table 2). Results of the phylogenetic analysis on deduced amino acids showed that IGF-II belongs to the super IGF family, with four receptor binding sites in domains A and B. In the comparison of IGF-II amino acid sequence of turbot with those ofParalichthy solivaceus(AF091454.1),Cynoglossus semilaevis(FJ608668.1), andEpinephelus coioides(AY552787.1), the four domains B, C, A, and D are more conservative while domain E and signal peptide region are less conservative (Fig.2). In the phylogenetic tree (Fig.3), bony fish clustered as a branch; wild boar, chicken, and claw toad form a branch; three sole fish (turbot, Japanese flounder, and half slippery tongue sole) form a small branch. Results of sequence comparison showed that the closest relative ofS. maximusis Japanese flounder (P. olivaceus), followed by half slippery tongue sole (C. semilaevis).
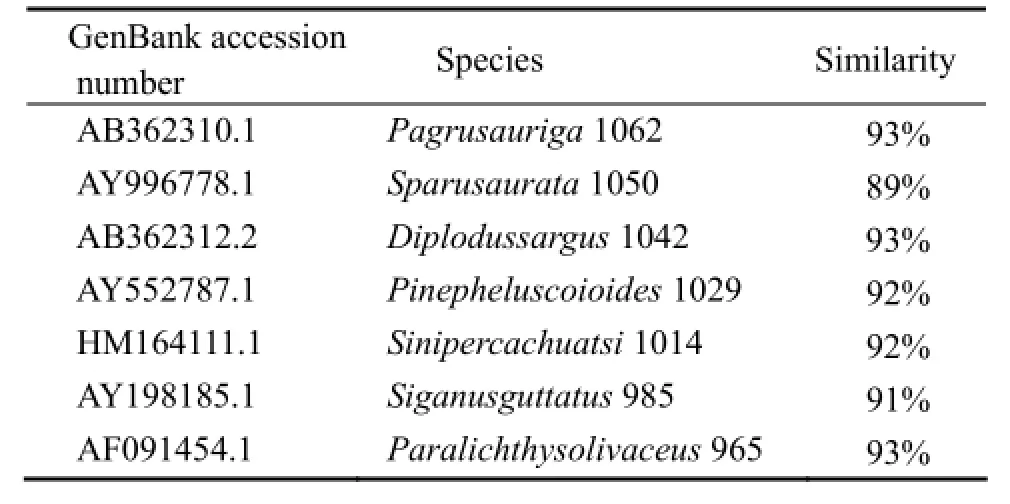
Table 2 Homologs ofScophthalmus maximusIGF-II

Fig.2 Alignment of amino acid sequences ofScophthalmus maximusIGF-II and those of other species. The identical, highly and less conserved amino acid residues are indicated by (*), (:), and (.), respectively; black backgrounds and white words indicate the 6 cysteine residues.
3.3 Tissue Distribution
The IGF-I gene was expressed in nine tissues of turbot except intestine (Fig.4). The IGF-I gene expression level was the highest in liver, followed by brain, spleen, kidney, heart, stomach, gills and skin, but hardly detected in muscle and ovary. On the other hand, the IGF-II gene was detected in all tissues (Fig.5). Its expression levels in spleen, brain, heart, stomach, liver, kidney, gills and intestine is higher than that in skin and muscle.
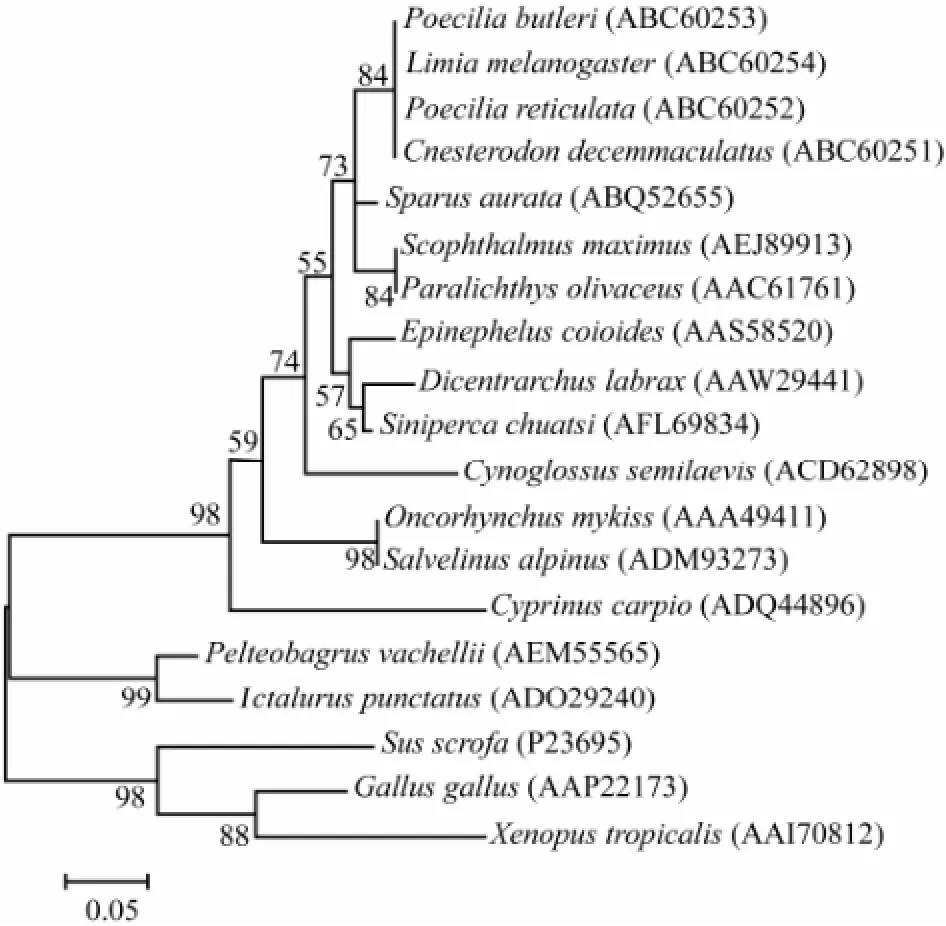
Fig.3 Phylogenetic tree constructed with amino acid sequences of IGF-II obtained in this study and those retrieved from GenBank. Phylogenetic analysis of IGF-II amino acid sequences inferred from the neighbor-joining method. GenBank accession numbers are in brackets after species names. Bootstrap values are indicated (1000 replicates).
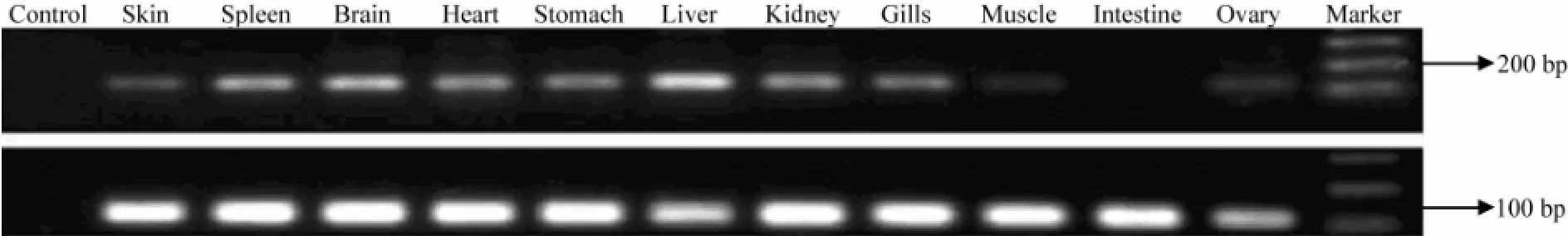
Fig.4 The tissue distribution of IGF-I gene transcript abundance inScophthalmus maximusassayed by RT-PCR. The integrity of RNA from each turbot tissue was detected by uniform amplification of 18S rRNA transcripts (lower line).
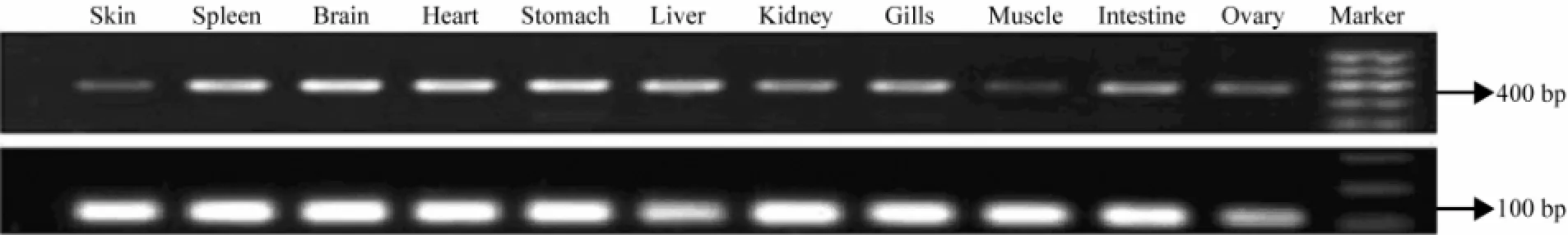
Fig.5 The tissue distribution of IGF-II gene transcript abundance inScophthalmus maximusassayed by RT-PCR. The integrity of RNA from each turbot tissue was detected by uniform amplification of 18S rRNA transcripts (lower line).
3.4 Gene Expression Profiling
Results of the RT-PCR assay (Fig.6) showed that during embryonic and larval development, the IGF-I mRNA abundance decreased from the unfertilized egg stage (0HPF) to the lowest level in the gastrula stage (18HPF), followed by a sharp increase at embryonic stage (80HPF) and a subsequent decrease at hatching stage (120HPF). The IGF-I gene mRNA abundance finally increased to the highest at post larva stage (25DPH). IGF-II gene showed a different expression pattern from IGF-I gene did, sharply increasing from unfertilized egg stage (0HPF) to gastrula stage (18HPF), then greatly decreased at embryonic stage (80HPF) and subsequently increased to the highest at post larva stage (25DPH).
4 Discussion
In this study, IGF-II gene was cloned from turbot liver. Additionally, the tissue distribution of IGFs and their expression patterns during embryonic and larval development of turbotS. maximuswere examined. Compared with other teleosts by amino acid sequence alignment, IGF-II ofS. maximusshared the greatest homology with that ofParalichthys olivaceus(91%). The high similarity between different IGF sequences suggested that these polypeptides are of importance to the growth and development of vertebrates. The results of amino acid sequence analysis revealed that IGF-II was divided into highly fragmented N-terminal signal peptide, conservative do-mains B, C, A, and D, and a less conservative domain E at C-terminus, with 6 conserved cysteine residues in the domains A and B. There are cysteine residues and IGF receptors binding protein amino acid residues in mammals’ IGF, which can explain why turbot has similar physiological function with mammals. This result is similar with that reported in IGF-II ofPsetta maxima(Duvalet al., 2002). Taking into account our previous finding of IGFBP in turbot (Huet al., 2012), we speculated that IGF-II could combine with 18 cysteine residues of IGFBP through the formation of disulfide bond.
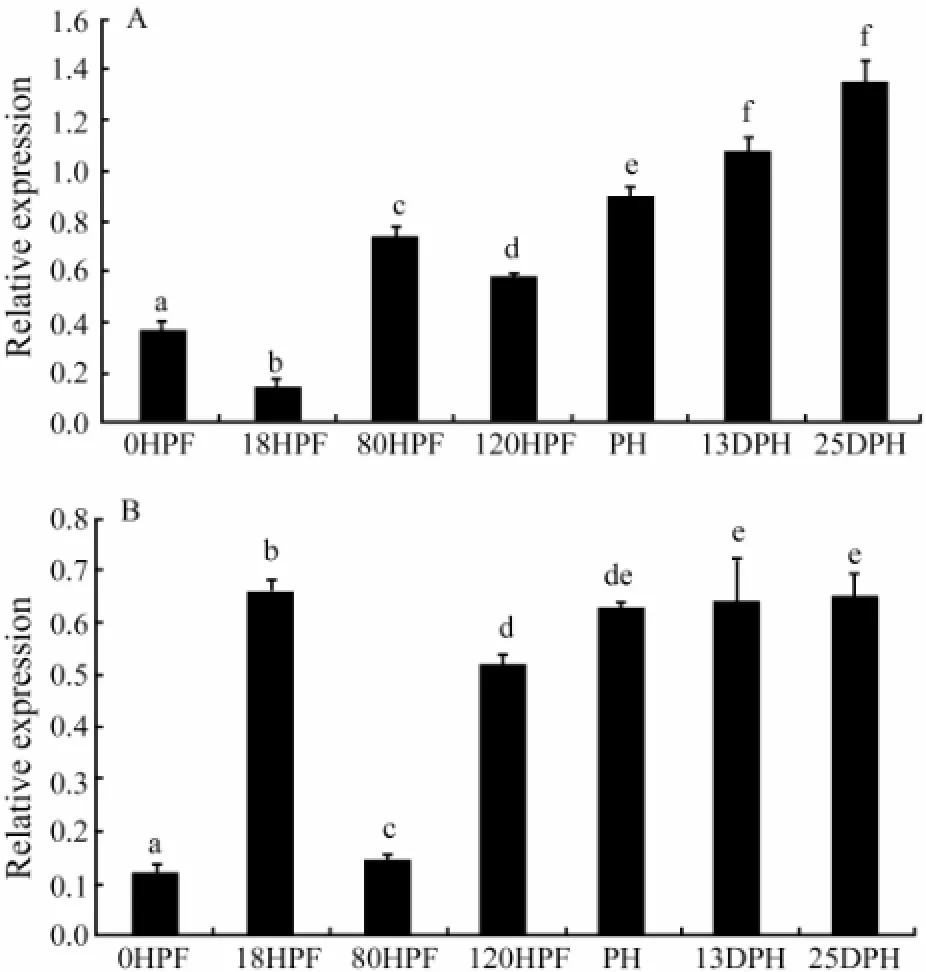
Fig.6 Expression of IGF-I (A) and IGF-II (B) genes during embryonic and larval developmental stages ofScophthalmus maximus. UE, unfertilized egg; PL, post larva; 13DPH, 13-day-post-hatching; PH, prophase of hatching; HS, hatching stage; ES, embryonic stage; GS, gastrula stage. The different letters indicate significant differences (P〈 0.05).
It is well known that IGF-I stimulates the growth, differentiation, proliferation and metabolism of cells (Langdahlet al., 1998). In the present study, the IGF-I gene expression level of turbot liver was significantly higher than those of other tissues. Similar results had been reported inSparus aurata(Duguayet al., 1996; Perrotet al., 1999),Cyprinus carpio(Tseet al., 2002; Vonget al., 2003),Oreochromis niloticus(Caelerset al., 2004),Soleasene galensis(Funeset al., 2006),Dicentrarchus labrax(Patrunoet al., 2008),Anguilla japonica(Moriyamaet al., 2006), andOreochromis hornorum(Gaoet al., 2012). The high expression of IGF-I in turbot liver indicated that IGF-I played a key role in the physiological function of liver. In addition, the nutritional conditions, ambient temperature, salinity and development state could affect IGF-I gene expression in liver (Moriyamaet al., 2006; Gaoet al., 2012). Although IGF-I is produced mainly in liver, it is also synthesized in a variety of tissues, including ovary, muscle, gills, kidney, stomach, heart, brain, spleen, and skin. This tissue distribution pattern of IGF-I was similar to that found in other fish species (Reinecke and Collet, 1998; Reineckeet al., 2006), which indicated that paracrine and autocrine actions of IGF-I were involved in organ-specific functions. The expression level of IGF-I gene in intestine was not detected in turbot, although it had been found low in other teleost species such as common carp (Vonget al., 2003) and rainbow trout (Greeneet al., 1997).
As compared to IGF-I, IGF-II showed a different expression pattern in turbot tissues. IGF-II gene was highly expressed in turbot spleen, brain, heart, stomach, kidney, gill and intestine except for liver. In Zanzibai tilapia, IGF-II gene was highly expressed in intestine, spleen, stomach, kidney and pituitary (Gaoet al., 2012), whereas inSparus aurata,Cyprinus carpio,Solea senegalensis, andDicentrarchus labrax, the IGF-II gene expression level was the highest in liver (Duguayet al., 1996; Tseet al., 2002; Funeset al., 2006; Terovaet al., 2007). The different expression patterns of IGF-II gene in various teleosts suggested that IGF gene expression was species-dependent. In addition, the expression of IGFs genes in turbot was weaker in muscle than in other tissues, despite IGFBPs were not detected in muscle in our previous study (Huet al., 2012). We speculated that the function of IGFs in muscle was controlled by other IGFBP family members. While the expression level of IGF-II gene in brain was higher than that of IGF-1, IGFBP-2 was highly expressed in brain of turbot (Huet al., 2012). It was possible that IGFBP-2 had higher affinity to IGF-II than to IGF-I (Binkertet al., 1989).
Studies showed that IGFs were synthesized at the early embryonic development stage. In Japanese flounder, IGF-I gene was expressed from the early embryonic stage and its expression was regulated at different embryonic stages. IGF-I mRNAs are hardly detected in early embryos, and the zygotic IGF-I gene transcripts gradually increased from the eye lens formation to pre-hatching stage (Zhanget al., 2012). IGF-I gene was detected in the embryos and larvae inSparus aurata,Oncorhynchus mykiss,Cyprinus carpio,Ictalurus punctatusand hybrid channel catfish (Ictalurus punctatus×I. furcatus) (Perrotet al., 1999; Greeneet al., 1999; Tseet al., 2002; Petersonet al., 2005). In addition, the IGF-1 gene transcription had been found in unfertilized eggs in gilthead seabream and rainbow trout (Perrotet al., 1999; Greeneet al., 1999). However, IGF-I mRNA was not detected inSiganus guttatusunfertilized eggs but was first expressed in larvae soon after hatching (Aysonet al., 2002). In the present study, the expression levels of IGF-I gene in unfertilized eggs, different embryonic development, and larvae stages were significantly different, implicating that IGF-I gene played an important regulatory role in embryo and larvae development.
In mammals, the circulating level of IGF-II gene was high before birth and declined afterwards (Moseset al., 1979). In the present study, results indicated that IGF-II gene was highly expressed after birth. Many studies demonstrated that IGF-II played a role not only in the embryonic period, but also at stages thereafter (Duguayet al., 1996; Perrotet al., 1999; Tseet al., 2002; Vonget al.,2003; Caelerset al., 2004; Funeset al., 2006; Patrunoet al., 2006; Hevroyet al., 2007; Radaelliet al., 2008). Furthermore, our study showed that the expression level of IGF-II gene in the gastrula stage was higher than those in other embryo and larvae development stages, consistent with our pervious findings of IGFBPs (Huet al., 2012). Together these results proved that there existed a relationship between IGF-II and IGFBPs in early embryo development. As a whole, the relative expression of IGF-II gene was less abundant than that of IGF-I gene in turbot, different from result in common carp. The IGF-II mRNA level was in fact higher than that of IGF-I in all tissues examined with an exception of liver (Tseet al., 2002). It is presumed that IGF-I played more important role than IGF-II for cell growth and tissue differentiation in the process of embryonic development in physiology during the early development of turbot (Chenet al., 2010). Additionally, the GH-dependent IGFs mRNA accumulation has been reported in teleosts (Sakamoto and Hirano, 1993), and our next work is to explore the function of the IGF system in GH release.
In conclusion, the full-length cDNA of IGF-II gene was isolated from turbot (S. maximus) liver in the present study. The result demonstrated that liver was the major tissue for IGF-I gene expression in turbot. In addition, we found that IGFs genes widely expressed in a variety of other tissues. To our knowledge, this study presented the first evidence of IGFs genes expression at the stages of embryonic and larval development, which showed that both IGFs genes played an important role at late embryonic stages. These findings contributed to understanding the functional mechanism of IGF system in teleosts.
Acknowledgement
This research was supported by the National Key Technologies R & D Program of China (Grant No. 2011BAD13B03).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D., 1990. Basic local alignment search tool.Journal of Molecular Biology, 215: 403-410.
Ayson, F. G., de Jesus, E. G., Moriyama, S., Hyodo, S., Funkenstein, B., Gertler, A., and Kawauchi, H., 2002. Differential expression of insulin-like growth factor I and II mRNAs during embryogenesis and early larval development in rabbitfish,Siganus guttatus.General and Comparative Endocrinology, 126 (2): 165-174.
Binkert, C., Landwehr, J., Mary, J. L., Schwander, J., and Heinrich, G., 1989. Cloning, sequence analysis and expression of a cDNA encoding a novel insulin-like growth factor binding protein (IGFBP-2).The EMBO Journal, 8 (9): 2497-2502.
Caelers, A., Berishvili, G., Meli, M. L., Eppler, E., and Reinecke, M., 2004. Establishment of a real-time RT-PCR for the determination of absolute amounts of IGF-I and II gene expression in liver and extrahepatic sites of the tilapia.General and Comparative Endocrinology, 137 (2): 196-204.
Chen, W., Wang, Y., Li, W. S., and Lin, H. R., 2010. Insulin-like growth factor binding protein-2 (IGFBP-2) in orange-spotted grouper, Epinepheluscoioides: Molecular characterization, expression pro fi les and regulation by 17β-estradiol in ovary.Comparative Biochemistry Physiology, 157 (4): 336-342.
Duguay, S. J., Zhang, J. L., Steiner, D. F., Funkenstein, B., and Chan, S. J., 1996. Developmental and tissue-regulated expression of, IGF-I and IGF-II mRNAs inSparus aurata.Society for Endocrinology, 16: 123-132.
Duval, H., Rousseau, K., Elies, G., Le Bail, P. Y., Dufour, S., Boeuf, G., and Boujard, D., 2002. Cloning, characterization, and comparative activity of turbot IGF-I and IGF-II.General and Comparative Endocrinology, 126 (3): 269-278.
Funes, V., Asensio, E., Ponce, M., Infante, C., Cañavate, J. P., and Manchado, M., 2006. Insulin-like growth factors I and II in the soleSolea senegalensise: cDNA cloning and quantitation of gene expression in tissues and during larval development.General and Comparative Endocrinology, 149 (2): 166-172.
Gao, F. Y., Lu, M. X., Huang, Z. H., Ping, Z. H., and Li, K. X., 2012. Cloning and sequence of two cDNAs encoding insulin-like growth factors I and II genes (IGF-I and IGF-II) of Zanzibar tilapia (Oreochromis hornorum) and their tissue distribution.Journal of Agricultural Biotechnology, 20 (2): 171-180.
Greene, M. W., and Chen, T. T., 1999. Quantitation of IGF-I, IGF-II and multiple insulin receptor family member messenger RNAs during embryonic development in rainbow trout.Molecular Reproduction and Development, 54: 348-361.
Hevroy, E. M., El-Mowafi, A., Taylor, R. G., Olsvik, P. A., Norberg, B., and Espe, M., 2007. Lysine intake affects gene expression of anabolic hormones in Atlantic salmon,Salmo solar.General and Comparative Endocrinology, 152 (1): 39-46.
Hu, J., Wen, H. S., Guan, J., Guan, S. G., He, F., Li, J. F., Shi, D., Ma, R. Q., Liu, M., Mu, W. J., and Zhang, Y. Q., 2012. Cloning of IGFBP-1,-2 and expression analysis during adult and early developmental stages inScophthalmus maximus.Acta Oceanologica Sinica, 34: 139-146 (in Chinese with English abstract).
Jones, J. I., and Clemmons, D. R., 1995. Insulin-like growth factors and their binding proteins: Biological actions.Endocrine Reviews, 16 (1): 33-34.
Lei, J. L., and Zhan, X. L., 2001. Experiment of rearingScophthalmus maximusLinnaeus by using deep well sea water in the farming plant.Modern Fish Information, 16 (3): 10-12 (in Chinese with English abstract).
Langdahl, B. L., Kassem, M., Møller, M. K., and Eriksen, E. F., 1998. The effects of IGF-I and IGF-II on proliferation and differentiation of human osteoblasts and interactions with growth hormone.European Journal of Clinical Investigation, 28: 176-183.
Moriyama, S., Yamaguchi, K., Takasawa, T., Chiba, H., and Kawauchi, H., 2006. Insulin-like growth factor I of Japanese eel,Anguilla japonica: cDNA cloning, tissue distribution, and expression after treatment with growth hormone and seawater acclimation.Fish Physiology and Biochemistry, 32 (3): 189-201.
Moses, A. C., Nissley, S. P., Passaman, J., and White, R. M., 1979. Further characterization of growth hormone-dependent somatomedin-binding proteins in rat serum and demonstration of somatomedin-binding proteins produced by rat liver cells in culture.Endocrinology, 104: 536-546.
Patruno, M., Sivieri, S., Poltronieri, C., Sacchetto, R., Maccatrozzo, L., Martinello, T., Funkenstein, B., and Radaelli, G.,2008. Real-time polymerase chain reaction, in situ hybridization and immunohistochemical localization of insulin-like growth factor-I and myostatin during development ofDicentrarchus labrax(Pisces: Osteichthyes).Cell Tissue Research, 331: 643-658.
Perrot, V., Moiseeva, E. B., Gozes, Y., Chan, S. J., Ingleton, P., and Funkenstein, B., 1999. Ontogeny of the insulin-like growth factor system (IGF-I, IGF-II, and IGF-1R) in gilthead seabream (Sparus aurata): Expression and cellular localization.General and Comparative Endocrinology, 116: 445-460.
Peterson, B. C., Bosworth, B. G., and Bilodeau, A. L., 2005. Differential gene expression of IGF-I, IGF-II, and toll-like receptors 3 and 5 during embryogenesis in hybrid (channel × blue) and channel catfish.Comparative Biochemistry Physiology, 141 (1): 42-47.
Radaelli, G., Poltronieri, C., Bertotto, D., Funkenstein, B., and Simontacchi, C., 2008. Cellular localization of insulin-like growth factor-II protein in the sea bass (Dicentrarchus labrax) from hatching to adult.Histology and Histopathology, 23 (5):523-530.
Reinecke, M., and Collet, C., 1998. The phylogeny of the insulin-like growth factors.International Review of Cytology, 183:1-94.
Reinecke, M., 2006.Insulin-Like Growth Factor I and II in Fish. Vol 1, Fish Endocr Inology. Reinecke, M.,et al., eds., Science Publishers, New Hampshire, Enfield, 87-130.
Rinderknecht, E., and Humbel, R. E., 1978. Primary structure of human insulin-like growth factor II.FEBS Letters, 89 (2):283-286.
Rosen, K. M., Wentworth, B. M., Rosenthal, N. L., and Villa-Komaroff, L., 1993. Specific, temporally regulated expression of the insulin-like growth factor II gene during muscle cell differentiation.Endocrinology, 133 (2): 474-481.
Saitou, N., and Nei, M., 1987. The neighbor-joining method: A new method for reconstructing phylogenetic trees.Molecular Biology and Evolution, 4 (4): 406-425.
Sakamoto, T., and Hirano, T., 1993. Expression of insulin-like growth factor I gene in osmoregulatory organs during seawater adaptation of the salmonid fish: Possible mode of osmoregulatory action of growth hormone.Proceedings of the National Academy of Sciences of the United State America, 90 (5): 1912-1916.
Sun, Y. Y., Zhang, Q. Q., Qi, J., Wang, Z. G., Chen, Y. J., Li, C. M., and Zhong, Q. W., 2008. Cloning and expression analysis of DMRT 1 gene inCynoglossus semilaevis.Jounal of WuhanUniversity (Natural Science Edition), 54 (2): 221-226.
Tamura, K., Dudley, J., Nei, M., and Kumar, S., 2007. MEGA4:Molecular evolutionary genetics analysis (MEGA) soft ware version 4.0.Molecular Biology Evolution, 24 (8): 1596-1599.
Terova, G., Rimoldi, S., Chini, V., Gomati, G., Bemardini, G., and Saroglia, M., 2007. Cloning and expression analysis of insulin-like growth factor I and II in liver and muscle of seabass (Dicentrarchus labrax,L.) during long term fasting and refeeding.Journal of Fish Biology, 70: 219-233.
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G., 1997. The CLUSTAL X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools.Nucleic Acids Research, 25 (24): 4876-4882.
Tse, M. C. L., Vong, Q. P., Cheng, C. H. K., and Chan, K. M., 2002. PCR-cloning and gene expression studies in common carp (Cyprinus carpio) insulin-like growth factor-II.Biochim et Biophysica Acta, 1575 (1-3): 63-74.
Upton, Z., Yandell, C. A., Degger, B. G., Chan, S. J., Moriyama, S. S., Francis, G. L., and Ballard, F. J., 1998. Evolution of insulin-like growth factor-I (IGF-I) action:in vitrocharacterization of vertebrate IGF-I proteins.Comparative Biochemistry Physiology, 121 (1): 35-41.
Vong, Q. P., Chan, K. M., and Cheng, C. H., 2003. Quantification of common carp (Cyprinus carpio) IGF-I and IGF-II mRNA by real-time PCR: Differential regulation of expression by GH.Journal of Endocrinology, 178: 513-521.
Werner, H., and Leroith, D., 1996. The role of the insulin-like growth factor system in human cancer.Advances in Cancer Research, 68: 183-223.
Yuan, X. N., Jiang, X. Y., Pu, J. W., Li, Z. R., and Zou, S. M., 2011. Functional conservation and divergence of duplicated insulin-like growth factor 2 genes in grass carp (Ctenopharyngodon idellus).Gene, 470 (1-2): 46-52.
Zapf, J., Froesch, E. R., and Schmid, C., 1999. Metabolic effects of IGFs. In:Contemporary Endocrinology 17: The IGF System-Molecular Biology, Physiology and Clinical Applications. Rosenfeld, R. G., and Roberts, C., eds., Humana Press, New Jersey, 788pp.
Zhang, J. L., Shi, Z. Y., Cheng, Q., and Zhai, W. Y., 2012. Expression patterns of IGF-I and its receptor genes during embryonic development of Japanese flounder.Journal of Fishery Sciences of China, 18: 1219-1225 (in Chinese with English abstract).
(Edited by Qiu Yantao)
(Received May 15, 2013; revised June 28, 2013; accepted December 20, 2014)
© Ocean University of China, Science Press and Spring-Verlag Berlin Heidelberg 2015
* Corresponding author. Tel: 0086-532-82031825 E-mail: wenhaishen@ouc.edu.cn
 Journal of Ocean University of China2015年2期
Journal of Ocean University of China2015年2期
- Journal of Ocean University of China的其它文章
- Impacts of the Two Types of El Niño on Pacific Tropical Cyclone Activity
- Numerical Simulation of Typhoon Muifa (2011) Using a Coupled Ocean-Atmosphere-Wave-Sediment Transport (COAWST) Modeling System
- Estimating the Turbulence Characteristics in the Bottom Boundary Layer of Monterey Canyon
- Composition and Origin of Ferromanganese Crusts from Equatorial Western Pacific Seamounts
- Hydroelastic Analysis of a Very Large Floating Structure Edged with a Pair of Submerged Horizontal Plates
- A Storm Surge Intensity Classification Based on Extreme Water Level and Concomitant Wave Height
