A Minireview of Marine Algal Virus - Coccolithoviruses
LIU Jingwen, XU Miaomiao, and ZHENG Tianling
1)College of Food and Bioengineering,Jimei University,Xiamen361021,P.R.China
2)Research Center of Food Microbiology and Enzyme Engineering Technology of Fujian Province,Xiamen361021,P.R.China
3)State Key Laboratory for Marine Environmental Sciences and Key Laboratory of the Ministry of Education for Coastal and Wetland Ecosystem,School of Life Sciences,Xiamen University,Xiamen361005,P.R.China
A Minireview of Marine Algal Virus - Coccolithoviruses
LIU Jingwen1),2), XU Miaomiao1),2), and ZHENG Tianling3),*
1)College of Food and Bioengineering,Jimei University,Xiamen361021,P.R.China
2)Research Center of Food Microbiology and Enzyme Engineering Technology of Fujian Province,Xiamen361021,P.R.China
3)State Key Laboratory for Marine Environmental Sciences and Key Laboratory of the Ministry of Education for Coastal and Wetland Ecosystem,School of Life Sciences,Xiamen University,Xiamen361005,P.R.China
Coccolithophorid is unicellular marine microalgae with a global distribution in temperate and sub-temperate oceanic regions and has the ability to produce ‘the coccoliths’. It is considered to be the second most productive calcifying organism on earth and becoming an important factor in the global carbonate cycle.Emiliania huxleyiis one of the only two bloom-forming coccolithophores and becomes a species crucial to the study of global biogeochemical cycles and climate modeling. Coccolithoviruse is a recently discovered group of viruses infecting the marine coccolithophoridE.huxleyi. They are a major cause of coccolithophore bloom termination, and DMSP concentration is increasing in the process of viral lysis. Phylogenetic evidences support that some genes are functional both inE.huxleyiand its virus (EhV). Horizontal gene transfer (HGT) of multiple functionally coupled enzymes occurs inE.huxleyiand its DNA virus EhV has been confirmed, which contributes to the diversification and adaptation of plankton in the oceans and also critically regulates virus-host infection by allowing viruses to control host metabolic pathways for their replication. Therefore, it is of particular interest to understand this host-virus interaction. On this issue, we have made a minireview of coccolithoviruses focusing on the basic characteristics, phylogenesis, horizontal gene transfer and the interaction between the host and its viruses, as well as its important role in global biogeochemical cycling.
coccolithoviruses; phylogenetic characteristics; virus-host interaction; horizontal gene transfer (HGT); global climate change
1 Introduction
Coccolithophorid (diameter of 3-10 μm) is unicellular, eukaryotic phytoplankton belonging to the class Prymesiophyceae in the division Haptophyta, which cells are covered with one or several layers of coccoliths that are composed of calcium carbonate. It’s widely distributed in offshore, coastal, oceanic waters, especially in the higher latitudes subpolar waters and its abundance can reach 5.0 × 106cells L-1(Holliganet al., 1983). Marine coccolithophorid has a unique biomineralization and high dimethyl sulphonio propionate (DMSP) emission capability, so the algae is a key species in regulating the global carbon cycle, sulfur biogeochemical cycle and climate change (Martínezet al., 2012; Ridgwell and Zeebe, 2005). Among the 250 kinds of coccolithophorid, onlyEmiliania huxleyiandGephyrocapsa oceanicaare bloom-forming species (Wilsonet al., 2002). The coccolithophoridE. huxleyiis one of the most successful eukaryotes in modern oceans. There are two phases in its haplodiploid life cycle exhibiting primarily different phenotypes.E.huxleyican escape viral attack by converting its life cycle from a diploid to haploid, and the diploid calcified phase forms mass blooms, which deeply influence global biogeochemical balance (Fradaet al., 2012).
Field and laboratory studies confirm that viral infection is an important factor in terminatingE. huxleyiblooms (Bratbaket al., 1993; Martínezet al., 2012). To date, 20E. huxleyiviral (EhVs) strains has been isolated from English channel and Norwegian fjords (Bratbaket al., 1993; Brussaardet al., 1996a; Castberget al., 2002; Schroederet al., 2002, Nissimovet al., 2014) and the genome of nine strains has been sequenced (Allen, 2006; Nissimovet al., 2011a, 2011b; Nissimovet al., 2012a, 2012b; Pagareteet al., 2013; Wilsonet al., 2005). The EhVs are lytic dsDNA viruses with genomes of approximately 400 kbp, belonging to a novel genus - Coccolithovirus within the familyPhycodnaviridae(Schroederet al., 2003).
Phylogenetic analysis shows that seven genes related to the sphingolipid biosynthesis pathway take place genes transfer betweenE. huxleyiand its specific EhVs, and EhVs also is the first confirmed eukaryotic phytoplankton-virus system involved in horizontal gene transfer of multiple functionally coupled enzymes (Monieret al., 2012). Interactions between marine viruses and phyto-plankton affect global biogeochemical cycles and climate. Therefore, it is of particular interest to understand the host-virus interaction.
2 Importance in Global Climate Change
Coccolithophores is a high producer of dimethylsulfoniopropionate (DMSP). Large viruses that infectE. huxleyiare largely responsible for the termination of these blooms. The demise ofE. huxleyiblooms also results in the release of dimethylsulfoniopropionate (DMSP) from the dying alga. WhenE. huxleyiare destroyed by virus, DMSP begin to crack and this behavior may benefits for fighting the destroying environment (Wolfeet al., 1997). DMS is formed from DMSP by enzymatic cleavage and it is the most abundant sulfide in the oceans accounting for over half of all nonanthropogenic gaseous sulfur input to the atmosphere (Bateset al., 1992; Van Etten, 2011). DMS oxidizes to produce methanesulphonic acid, SO2and sulphuric acid, promotes the acidity of aerosol, which is a main source of cloud-condensation nuclei and may have impact on climate regulation (Vogtet al., 2008; Arnoldet al., 2013).
E.huxleyialso plays a remarkable role in the marine carbon cycle. A highly comprehensive overview about the role ofE.huxleyiin the ocean and climate has been reported (Westbroeket al,. 1993). Its daedal calcite coccoliths account for about 1/3 of the total marine CaCO3production (Vardiet al., 2012). Coccolithophores can use the CO2released in the calcification reaction for photosynthesis (Mackinderet al., 2009). The production of calcium carbonate reduces the alkalinity of ocean surface, which results in the CO2release back into the atmosphere. Consequently, large blooms of coccolithophores may contribute to global warming in a short time (Bateset al., 1996; Bachet al., 2013). However, for a long time period coccolithophores are more likely to promote an overall decline in atmospheric CO2concentrations. Calcium carbonate as the form of coccoliths sinks to the bottom of the ocean and becomes part of sediment, therefore, coccolithophores provide a sink for emitted carbon, regulating the effects of greenhouse gas emissions (Marsh, 2003). Viral infection of coccolithophores affects coccoliths formation and sinking, so the formation, shedding and deposition of coccoliths will surely influence the concentration of CO2both in the atmosphere and in the oceans while CaCO3is deposited on the ocean floor and participates in geological evolution (Rickabyet al., 2007; Van Etten, 2011).
3 Viral Control of E. huxleyi Blooms
Viral-mediated mortality can have impact on both algal species succession and intraspecies succession. Viruses affect the fluxes of energy, nutrients, and organic matter indirectly by lysis of the hosts’ cells, particularly during the algal bloom (Bratbaket al., 1996). Therefore viruses can control blooms by reducing host populations, or by keeping them from reaching high levels (Brussaard, 2004).
Viruses are essentially connected to the decline ofE.huxleyiblooms. However, the mechanisms of viral infection are poorly understood. One of the main mechanisms ofE. huxleyiblooms extinction is considered to be viral lysis (Vardiet al., 2012). For example, EhV86 produces a glycosphingolipid (GSL) that triggers programmed cell death (PCD) with corresponding activation of an algal metacaspase inE. huxleyi(Bidleet al., 2007; Vardiet al., 2009). EhV86 particularly has some unusual CDSs (Coding sequence), involving some genes encoding sphingolipid synthesis enzymes (Monieret al., 2009). This biosynthetic pathway appears to function during lytic infection and PCD. Virus can limit host population size, thus preventing the development of blooms. ForE.huxleyi, in the beginning of infection, virus can not prevent the accumulation of hosts’ cells because of its lower concentration, whereas the infection rates potentiallly increase with the increased concentration of host cells. Eventually, the host abundance exceeds a threshold, leading to rapid spread of infections, and a relatively quick decrease of the bloom occurs (Bratbaket al., 1996)
4 The Abundance of EhVs
Viruses are the most abundant up to 107to 108particles mL-1and genetically diverse biological entities in the sea (Suttle, 2005). Many of the algal virus strains show highly specific host ranges, so if a bloom is dominated by a specific algal strain, the virus specific to that strain is most abundant. So EhVs can be found in these region whereE.huxleyiexists. The abundance of EhVs in natural seawater can typically attain to 107VLP (viral-like particle) mL-1during bloom and 108to 109VLP mL-1under laboratory culture (Schroederet al., 2003). In the different stages ofE.huxleyiblooms with different abun- dance, the number can reach 3.7×107cells L-1during the termination phase of theE.huxleyibloom (Sorensenet al., 2009).
5 Life History of EhVs
Infection of algal viruses is highly host-specific such as the prototype virus EhV-86 infects onlyE.huxleyistrains (Schroederet al., 2003). EhV is a lytic double-stranded DNA virus with a genome size of about 415 kilo-base pairs, which has a life history of 7000 years (Coolen, 2011). The viral particle is an icosahedron with a diameter of 160-180 nm, and consists of envelope, coat protein and genome. Its capsid is enveloped by a lipid membrane (Mackindeet al., 2009). It contains nine proteins ranging from 10 to 140 kDa and the major capsid protein (MCP) weighs about 54 kDa. During infection, the virus coat protein-adsorbed on the surface of host cell or binds to the coccoliths, then the virus invades its host cell with its nucleoprotein particles together. This makes a distance of several nanometers between the bound particle and the cell surface (Duniganet al., 2006). This infers that virus infection will lead toE.huxleyicells collapse and the coccoliths dropping. The latent period of EhV is 12-14 hand the burst size is 400-1000 viral particles per cell; the virus start to assemble progeny virions in the host cytoplasm after 24 hours of infection (Castberget al., 2002). Based on the research of the EhV-86, it has been found that nascent coccolithoviruses may be released from their host by utilizing cellular degradative pathways although it lacks genes encoding cell wall degrading enzymes, (Wilsonet al., 2005).
6 Phylogenetic Characteristics of EhVs
Traditionally, the DNA polymerase gene is used to study the diversity and phylogeny of phycodnaviruses (Chenet al., 1996; Schroederet al., 2003). In recent years the gene encoding the major capsid protein (MCP) has also been used as an alternative marker, capable of distinguishing phylogenetic differences on a strain level (Larsenet al., 2008; Pagareteet al., 2009; Roweet al., 2011). The diversity of EhV is mainly reflected in its host range. According to their host range, 14 virus strains can be clustered into three groups based on Bray-Curtis similarity, as EhV-163 relatively narrow host ranges and lack ability to infect the same host strains of other EhV being a group solely (Fig.1) (Allenet al., 2006).
The DNApolgene fragment sequences of the EhV isolates can not differentiate between the isolates, while virus isolates are verified as being different from each other when the fragments from a putative MCP gene are compared. Thus, it appears that the DNApolgene in EhVs are evolutionarily more conserved than the major capsid protein gene. The phylogenetic analysis based on MCP could show the phenotypic diversity in coccolithoviruses as far as host range. The infection between virus and cells starts up by virus MCP and cell membrane conjugation. The difference of virus infection in specificity and susceptibility is due to the viral capsid. Therefore, such structural differences may also contribute to the viral successfully infecting and attaching to the host cells. The way EhV infect hosts is different from any other algae viruses but is similar to animals virus infection. It is reported that EhV-86 enters its host via either an endocytotic or an envelope fusion mechanism in which an intact nucleoprotein core is encapsulated by MCP, which is different from that employed by other algal NCLDVs (nucleocytoplasmic large DNA viruses) (Mackinderet al., 2009). Based on this, EhV-MCP could be expected to be as a marker for the detection of EhVs infectied host cells (Liuet al., 2013).
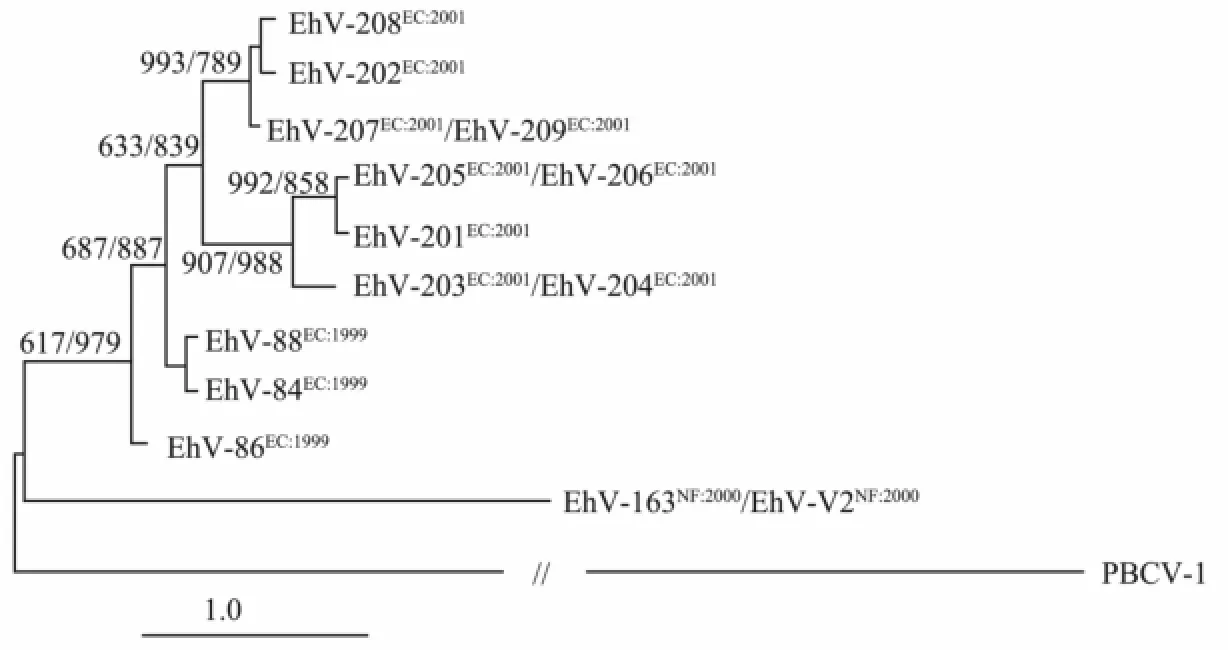
Fig.1 Phylogenetic inference tree based on a distance matrix algorithm between the sequences of DNA polymerase and MCP gene for members of the coccolithovirus family.Paramecium bursariaChlorella virus 1 (PBCV-1) is used as an outgroup (Neighbor, in PHYLIP version 3.6b). Numbers at junctions indicate bootstrap values retrieved from 1000 replicates for both the neighbour-joining and parsimony analyses. The bar depicts one base substitution per 100 nucleotides (Allenet al., 2006).
The evolutionary arms race between virus and the host is something that the virus must consider and fit to; and this might explain the variability of the gene among strains. Diversity of EhV strains researched by microarray shows that EhV-207, EhV-209, EhV-163 and EhV-V2 have identical MCP and DNA pol gene fragment sequences, but there is a significant difference on the coding core-specific clusters (Locus-specific groups) CDSs coupled with the virus proliferation. The difference is considered to have an important impact on EhV virulence and host range selection (Allenet al., 2006). These studies provide a fact that EhV are unique in NCLDVs family even among all known viruses. This suggests that EhV probably has an unique evolutionary history and lifestyle. The obvious difference in evolutionary divergence rates of DNA pol and MCP genes is interesting and implies that horizontal transfer of genetic material between different coccolithovirus genomes may be popular in the natural environment (Nissimovet al., 2011a).
SPT is the first and rate-limiting enzyme in thede novosphingolipid biosynthesis pathway and homologues are encoded by both the virus and host genomes (Hanet al., 2006; Monieret al., 2009; Wilsonet al., 2005). It has been considered to be involved in the mass termination of coccolithophore bloomsviathe propagation of programmed cell death (PCD) of its host (Bidleet al., 2011; Vardiet al., 2009, 2012). Recently, a way combining the‘standard’ markers MCP with metabolically relevant markers SPT is used for phylogeny and functionality in a study to assess both spatial and temporal variability (Nissimovet al., 2013). Nine new coccolithovirus genotypes with the majority of MCP sequences and four new SPTgene variations were discovered, which improves the current understanding of the classification of these viruses and their distribution, and offers an insight into their functional biodiversity and ecological relevance (Nissimovet al., 2013)
7 Genome of of EhVs
7.1 Genome Structure of of EhVs
EhV-86, the model virus, isolated in 1999 from the English Channel has been fully sequenced, in which 472 CDSs were identified (Wilsonet al., 2002, 2005), while a further eight strains (EhV-163, EhV-201, EhV-202, EhV-203, EhV-207, EhV-208, EhV-84, EhV-88 and EhV-99B1) have been partially sequenced (Allenet al., 2006; Nissimovet al., 2011a, 2011b; Nissimovet al., 2012a, 2012b; Pagareteet al., 2012). The sequence analysis of ten EhVs revealed that all of them contain dsDNA genomes ranging from 390 to 420 kb, which belong to the nuclear-cytoplasmic large double-stranded DNA virus (NCLDV) groups.
EhV genomes have many similar characteristics: 1) the relatively high and stable G+C % content, from 39.87% (EhV-BB91) to 40.49% (EhV-207); 2) All of them contain more than 450 predicted protein coding sequences (CDSs); 3) 35 insertion/deletion events could be identified between EhV-99B1 and EhV-86, including CDSs whose protein products are predicted to be coupled with activities such as endonuclease, phosphate permease and transposase activities, as well as a C2H2type zinc finger protein, DNA-binding protein and 12 putative membrane proteins (Pagareteet al., 2012). However, both phosphate permease and endonuclease belong to conservative members of the nucleic acid hydrolase family. EhV-86 genome encodes 5 tRNA genes: Leu, Arg, Gln, Asn, and Ile (Wilsonet al., 2005). By comparing EhV-86 genome and EhV-99B1 genome it is found that EhV-99B1 genome also encodes 5 tRNAs, but the tRNA gene for leucine is replaced by another tRNA gene with the lysine anti-codon. In a sense, the presence of different tRNAs in these two EhV genomes could either be directly linked to the codon usage inherent to each viral genome, or, if they have different host ranges, this could be due to different codon-selective pressures imposed by the translational biases of their respective hosts. The precise function for gene replacement remains unknown, whereas the function of similar genes to replace likely reflects the different properties of EhV adaptation to the environment (Nissimovet al., 2012a). Except for gene replacement phenomenon, at least 16 kinds of CDSs, with the same function, coded by EhV-99B1 and EhV-86, have a clear base insertion and deletion. These changes result in changes in protein structure which could explain phenotypic difference to be shown by EhV. These studies indicate that EhV strains not only have a very close correlation, also have a certain variability, which probably reflects the EhV evolutionary adaptation of different hosts to natural environment.
7.2 Functional Genes
EhVs contain six RNA polymerase subunits and many promoters, indicating that this virus encodes its own transcription machinery (Wilsonet al., 2005). In addition, it is amazing that EhVs encode a remarkable array of proteins and even metabolic pathways, as well as properties that are provided with the function of the host. However, the functions of more than 50% of viral CDSs function are still unknown. Only a few unusual virus- encoded properties are mentioned due to space limitations and a large proportion of CDSs show no similarity to proteins in the public databases (Wilsonet al., 2005; Van Eet al., 2011). One key synthase gene predicted to encode serine palmitoyltransferase (SPT), the first and committed enzyme in de novo sphingolipid biosynthesis pathway (SBP), was discovered in EhV-86 genome (Wilsonet al., 2005). SPT presented in EhV86 is considered closely related to the host ceramide synthesis and host programmed cell death (Sutterwalaet al., 2007; Vardiet al., 2009). For a functional identification, the EhV99B1-SPT subunit-LCB2 gene was expressed inEscherichia coli, which resulted in significant accumulation of new sphingolipid inE.colicells (Liuet al., 2012). Another recently found gene in EhV99B1 genome is ‘inteins’, which are selfish DNA elements found within coding regions of host proteins (Allenet al., 2011; Pagareteet al., 2012). The newly identified coccolithovirus intein is predicted to have both self-splicing and homing endonuclease of mediating their transfer and insertion into unoccupied integration sites, thus facilitating horizontal gene transfer (HGT) (Allenet al., 2011; Nagasakiet al., 2005). Thioredoxin (Trx)-like gene discovered in EhV is identifiable as a new member of thioredoxin family. Trx is a great important component of the thiol-reducing system and regulates various cellular functions (redox regulation) and environment stress (Masutaniet al., 2005). Recombinant EhV99B1-Trx shows antioxidant function and significant disulfide bond reduction effect on insulin molecule (Zhanget al., 2010; Caiet al., 2012).
8 The Relationship Between EhV and Host
Viral infection affects various metabolism of host cells such as photosynthesis, transcription and translation, carbohydrate and lipid metabolism (particularly glycolysis), and signal transduction (Kegelet al., 2010). Marine viruses including eukaryotic viruses, phage and gene transfer agents (GTAs), have dramatic effects on host cells through genetic materials transform. When a phage infects its host cell, it can transfer genetic material by horizontal gene transfer or accessory genes expression, which can make host have a broader niche (Rohwer and Thurber, 2009). Moreover, small viral-like particles known as GTAs can transfer genes between marine organisms. In the ‘Red Queen’ effect, the virus and host cell are locked in an evolutionary ‘arms race’, so that they could keep to co-evolve mechanisms of resistance to each other untilultimately the host cell is lysed. InE.huxleyi, the life cycle comprises two main forms: the diploid (2N), nonmotile, coccolith-bearing phase and the haploid (N) flagellated phase (Bratbaket al., 1996). EhVs can infect and lyse diploid-phase cells and is very relevant in the regulation of populations and blooms termination. The haploid phase ofE.huxleyiis unrecognizable and thus resistant to EhVs that kill the diploid phase. In the ‘Cheshire Cat’hypothesis, the coccolithophorid simply changes its life cycle from a diploid, non-mobile stage to a motile, haploid stage, thus escaping the virus (Fig.2) (Fradaet al., 2008; Rohwer and Thurber, 2009). This ‘Cheshire Cat’ecological dynamics reduces pathogen pressure during host evolution and it can be considered an opposite force to a classic ‘Red Queen’ coevolutionary arms race. InE.huxleyi, this can explain why the selective balance tend to the boom-and-bust situation using optimization of both growth rates of diploidE. huxleyicells and infectivity of EhVs (Fradaet al., 2008). In addition, the expression of cell cycle dependent proteins (cyclins) can be altered by EhV infection, thus inhibiting the progression of host-cell cycle, which indicates that algal virus infection selectively activates/inactivates certain components of the cell cycle with the aim to establish a more efficient environment for their gene expression and DNA replication (Liuet al., 2011).
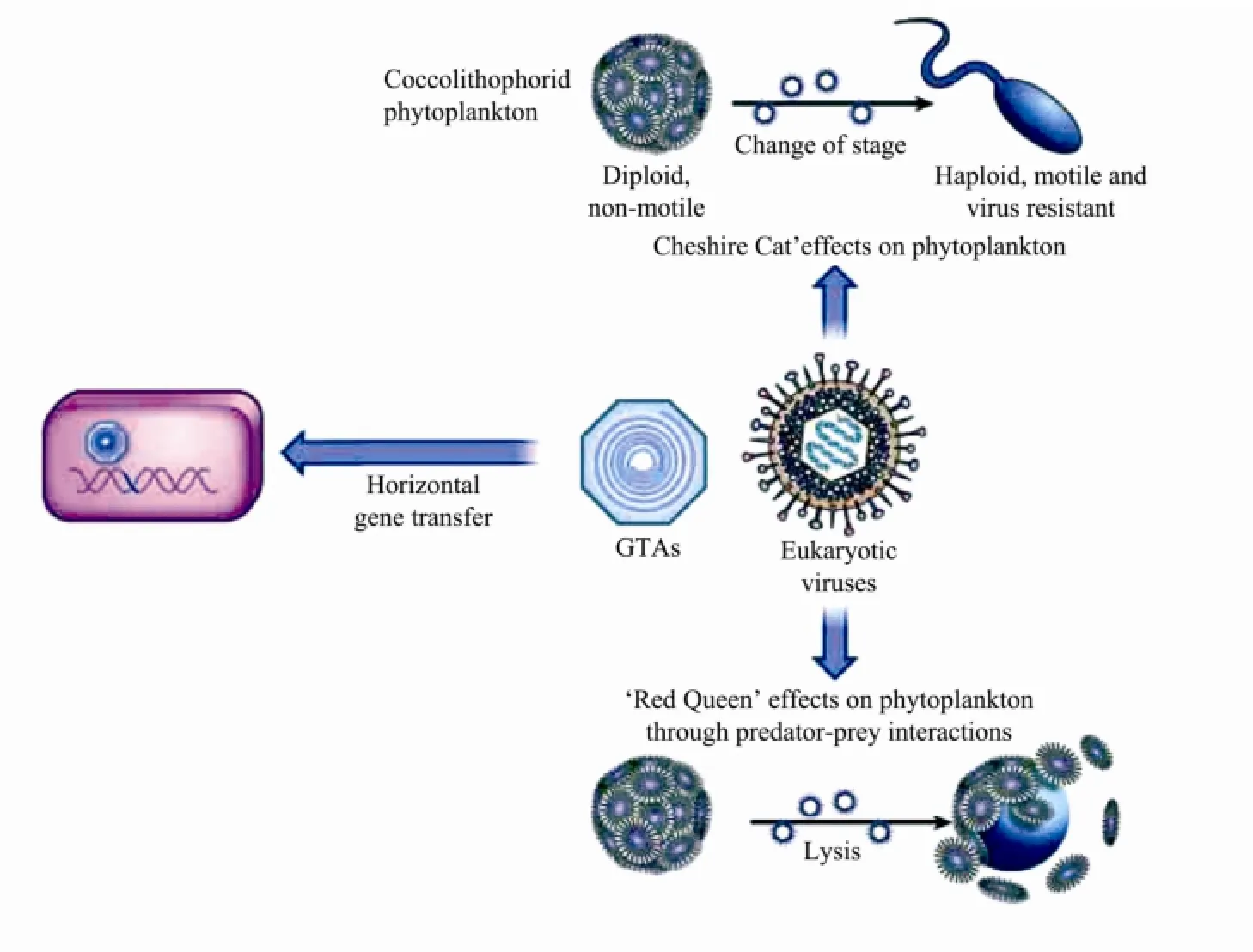
Fig.2 Effects ofE. huxleyivirus on its hosts (Rohwer and Thurber, 2009).
9 Horizontal Gene Transfer of the EhVs
Horizontal gene transfer (HGT) refers to the process during which an organism acquires genetic material from another organism but not being the offspring of that organism, and then integrates these genes into its own genomes and expresses specific functions. HGT contributes to the diversification and adaptation of plankton in the oceans and also critically regulates virus-host infection by allowing viruses to control host metabolic pathways for their replication (Bidle and Vardi, 2011). Recently, increasing studies show the possible cases of gene transfers between cyanobacteria and phages, which might have played prominent roles in the co-evolution of cyanobacteria/phage systems (Zhaxybayevaet al., 2007). Gene transfer agents (GATs), viral-like particles produced by α-Proteobacteria result in horizontal gene transfer in coastal and oceanic environments (McDanielet al., 2010).
From genome sequence of EhVs, it is surprising that EhVs encode an array of proteins and even metabolic pathways, as well as characteristics that are provided with the function of the host (Bidle and Vardi, 2011; Monieret al., 2012; Nissimovet al., 2011a, 2011b; Vardiaet al., 2012). Seven genes are predicted coding key catalytic enzyme of SBP in EhV genomes. These genes are only present in EhV uniquely suggests that HGT of these functionally coupled enzyme gene may exist between ancestral virus and eukaryotic host (Monieret al., 2009). The dendrographic structure (branching topology) constructed by the seven genes shared by EhV andE. huxleyicontains the fundamental information about the phylogenetic origin in which the EhV/E. huxleyisequence groups for these genes might be generally evolved fromE. huxleyiwithin eukaryotes, except for the Aco1-like FADs (Monieret al., 2009). These results indicate that the seven sphingolipid biosynthesis genes have been horizontally transferred between viral lineages and the eukaryotic, leading toE. huxleyiand EhVs, respectively (Monieret al., 2009).
However, the direction of horizontal gene transfer be-tween virus and host remains uncertain. One theory is that viral SPB genes come from their hosts based on ‘cell source’ theory and ‘gene robbers’ theory (Forterre and Prangishvili, 2009). Another theory is that SBP genes of virus are horizontally transferred to the host based on‘eukaryogenesis’ theory (Bell, 2001; Takemura, 2002) and ‘ancient Virus’ theory (Wilsonet al., 2002). The hypothesis conforms to the evolutionary distance between EhV and host. If the hypothesis is right, the EhV genome characteristics could be found in most eukaryotic cells. Only the host and the virus have a certain homogeneity actually. So the existing evidence can prove horizontal gene transfer occurred between EhV and its host, but can not explain the specific direction of horizontal gene transfer.
10 Conclusions
Coccolithoviridae are a group of viruses which infect the marine coccolithophoridE. huxleyi, a globally distributed bloom-forming marine microalga. Viral infection can terminateE. huxleyiblooms, and facilitate the release of DMSP in the process of viral lytic. Although viral infections are usually specific to certain host strains in a species, lytic viral infections nevertheless affect a large proportion of algae and have a global impact, for example, in the termination of blooms. Resistance to viruses is thus subject to strong selection, but little is known about its mechanism. The interactions between viruses and phytoplankton in the oceans influence global biogeochemical cycles and climate. EhV are unique in NCLDVs family, even all known viruses. This implies that EhV probably has a unique evolutionary history and a unique lifestyle.
The genome of EhVs analysis of protein sequences predicts novel functions and many of them have no current matches in the sequence databases. Many of the CDSs show high conservation with those in EhV-86 model virus, while a few of highly variable CDSs indicate roles in evolutionary adaptation to their hosts. Phylogenetic evidence confirms the transfer of seven genes involved in the sphingolipid biosynthesis pathway between the eukaryotic microalgaE. huxleyiand its large DNA virus EhV. Patterns of protein and gene sequence conservation support that these genes are functional both inE. huxleyiand EhV. Lateral gene transfer in eukaryotic phytoplankton/virus systems has been shown by comparisons between their complete genomes and must play an important role in coevolution in the microbial world. Advances in bioinformatics and the possibility of amplifying complete genomes from single cells promise to revolutionize analyses of viral genomes from environmental samples.
Acknowledgements
This work was funded by the Chinese Public Science and Technology Research Funds Projects of Ocean (No. 201305027) and the National Natural Science Foundation of China (Nos. 40930847, 41376119), Funds of China Southern Oceano- graphic Research Center (No. 14GZP71NF35) and Funds of Provincial Key Laboratory of Food Microbiology and Enzyme Engineering (No. M20140910).
Allen, M. J., Lanzén, A., and Bratbak, G., 2011. Characterisation of the coccolithovirus intein.Marine Genomics, 4 (1): 1-7.
Allen, M. J., Schroeder, D. C., Donkin, A., Crawfurd, K. J., and Wilson, W. H., 2006. Genome comparison of two Coccolithoviruses.Virology Journal, 3: 15, DOI: 10.1186/1743-422X-3-15.
Arnold, H. E., Kerrison, P., and Steinke, M., 2013. Interacting effects of ocean acidification and warming on growth and DMS-production in the haptophyte coccolithophoreEmiliania huxleyi.Global Change Biology, 19 (4): 1007-1016.
Bach, L. T., Mackinder, L. C., Schulz, K. G., Wheeler, G., Schroeder, D. C., Brownlee, C., and Riebesell, U., 2013. Dissecting the impact of CO2and pH on the mechanisms of photosynthesis and calcification in the coccolithophoreEmiliania huxleyi.New Phytologist, 199 (1): 121-134.
Bates, N. R., Michaels, A. F., and Knap, A. H., 1996. Alkalinity changes in the Sargasso Sea; geochemical evidence of calcification?Marine Chemistry, 51: 347-358.
Bates, T. S., Lamb, B. K., Guenther, A., Dignon, A., and Stoiberet, R. E., 1992. Sulfur emissions to the atmosphere from natural sources.Journal of Atmospheric Chemistry, 14:315-337.
Bell, P. J., 2001. Viral eukaryogenesis: Was the ancestor of the nucleus a complex DNA virus?Journal of Molecular Evolution, 53 (3): 251-256.
Bidle, K. D., 2007. Viral activation and recruitment of metacaspases in the unicellular coccolithophore,Emiliania huxleyi.PNAS Proceedings of the National Academy of Sciences of the United States of America, 104 (14): 6049-6054.
Bidle, K. D., and Vardi, A., 2011. A chemical arms race at sea mediates algal host-virus interactions.Current Opinion in Microbiology, 14 (4): 449-457.
Bratbak, G., Egge, J. K., and Heldal, M., 1993. Viral mortality of the marine algaEmiliania huxleyi(Haptophyceae) and termination of algal blooms.Marine Ecology-Progress Series, 93 (1-2): 39-48.
Bratbak, G., Wilson, W., and Heldal, M., 1996. Viral control ofEmiliania huxleyiblooms?Journal of Marine Systems, 9 (1-2): 75-81.
Brussaard, C. P. D., Kempers, R. S., Kop, A. J., Riegman, R., and Heldal, M., 1996a. Virus-like particles in a summer bloom ofEmiliania huxleyiin the North Sea.Aquatic Microbiology Ecology, 10: 105-113.
Brussaard, C. P. D., 2004. Viral control of phytoplankton populationsa Review.Journal of Eukaryotic Microbiology, 51 (2):125-138.
Cai, Y. Q., Zhang, Z. L., Luo, B. B., and Liu, J. W., 2012. Expression and activity analysis of thioredoxin from marine coccolithophoridEmiliania huxleyiinPichia pastoris.Oceanologia et Limnologia Sinica, 43 (5): 905-910.
Castberg, T., Thyrhaug, R., Larsen, A., Sandaa, R. A., and Heldal, M .V., 2002. Isolation and characterization of a virus that infectsEmiliania huxleyi(Haptophyta).Journal of Phycology, 38 (4): 767-774.
Chen, F., Suttle, C. A., and Short, S. M., 1996. Genetic diversity in marine algal virus communities as revealed by sequenceanalysis of DNA polymerase genes.Applied Environment Microbiology, 62: 2869-2874.
Conte, M., van der Wal, P., Veldhuis, M., Knappertsbusch, M., Stefels, J., Brownlee, C., van Bleijswijk, J., Westbroek, P., Young, J., Fernandez, E., Brown, C. W., Jordan, R., Egge, J., and Brummer, G. J., 1993. A model system approach to biological climate forcing. The example ofEmiliania huxleyi.Global and Planetary Change, 8 (1-2): 27-46.
Coolen, M. J. L., 2011. 7000 Years ofEmiliania huxleyiviruses in the Black Sea.Science, 333 (6041): 451-452.
Dunigan, D. D., Fitzgerald, L. A., and Van Etten, J. L., 2006. Phycodnaviruses: A peek at genetic diversity.Virus Research, 117 (1): 119-132.
Forterre, P., and Prangishvili, D., 2009. The origin of viruses.Research in Microbiology, 160 (7): 466-472.
Frada, M. J., Bidle, K. D., Probert, I., and de Vargas, C., 2012. In situ survey of life cycle phases of the coccolithophoreEmiliania huxleyi(Haptophyta).Environmental Microbiology, 14 (6): 1558-1569.
Frada, M., Probert, I., Allen, M. J., Wilson, W. H., and de Vargas, C., 2008. The ‘Cheshire Cat’ escape strategy of the coccolithophoreEmiliania huxleyiin response to viral infection.Proceedings of the National Academy of Sciences of the United States of America, 105 (41): 15944-15949.
Han, G., Gable, K., Yan, L., Allen, M. J., Wilsonm W. H., Moitra, P., Harmon, J. M., and Dunn, T. M., 2006. Expression of a novel marine viral single chain serine palmitoyltransferase and construction of yeast and mammalian single-chain chimera.Journal of Biology Chemistry, 281: 39935-39942.
Hill, R. W., White, B. A., Cottrell, M. T., and Dacey, J. W. H., 1998. Virus-mediated total release of dimethylsulfoniopropionate from marine phytoplankton: A potential climate process.Aquatic Microbial Ecology,14: 1-6.
Holligan, P. M., Viollier, M., Harbour, D. S., Camus, P., and Champagne-Philippe, M., 1983. Satellite and ship studies of coccolithophore production along a continental shelf edge.Nature, 304 (5924): 339-342.
Kegel, J. U., Blaxter, M., Allen, M. J., Michael, J., Metfies, K., Wilson, W. H., and Valentin, K., 2010. Transcriptional host-virus interaction ofEmiliania huxleyi(Haptophyceae) and EhV-86 deduced from combined analysis of expressed sequence tags and microarrays.European Journal of Phycology, 45 (1): 1-12.
Liu, J. W., Zheng, T. L., Bratbak, G., and Thyrhaug, R., 2011. Virus infection disturbs cyclin expression, leading to cell cycle arrest in the unicellular marine algaeEmiliania huxleyiandChrysochromulina ericina.African Journal of Microbiology Research, 5 (14): 1801-1807.
Liu, J. W., Zhang, Z. L., Liu, X. H., Cai, Y. Q., and Cai, H. N., 2013. Cloning, expression of the major capsid protein gene from marine algaeEmiliania huxleyivirus and the possible use in detection of virus infection.Microbiology Research, 4 (1): 21-25.
Liu, X. H., Zheng, T. L., Cai, Y. X., and Liu, J. W., 2012. Cloning, expression and characterization of serine palmitoyltransferase (SPT)-like gene subunit (LCB2) from marineEmiliania huxleyivirus (Coccolithovirus).Acta Oceanologica Sinica, 31 (6): 127-138.
Mackinder, L. C. M., Worthy, C. A., Biggi, G., Hall, M., Ryan, K. P., Varsani, A., Harper, G. M., Wilson, W. H., Brownlee, C., and Schroeder, D. C., 2009. A unicellular algal virus,Emiliania huxleyivirus 86, exploits an animal-like infection strategy.Journal of General Virology, 90 (9): 2306-2316.
Marsh, M. E., 2003. Regulation of CaCO3formation in coccolithophores.Comparative Biochemistry Physiology B: Biochemistry Molecular Biology, 136: 743-754.
Martínez, J. M., Schroeder, D. C., and Wilson, W. H., 2012. Dynamics and genotypic composition ofEmiliania huxleyiand their co-occurring viruses during a coccolithophore bloom in the North Sea.FEMS Microbiology Ecology, 81 (2):315-323.
Masutani, H., Ueda, S, and Yodoi, J., 2005. The thioredoxin system in retroviral infection and apoptosis.Cell Death and Differentiation, 12 (1): 991-998.
McDaniel, L. D., Young, E., Delaney, J., Ruhnau, F., Ritchie, K. B., and Paul, J. H., 2010. High frequency of horizontal gene transfer in the oceans.Science, 330 (6000): 0036-8075.
Monier, A., Pagarete, A., de Vargas, C., Allen, M. J., Read, B., Claverie, J. M., and Ogata, H., 2009. Horizontal gene transfer of an entire metabolic pathway between a eukaryotic alga and its DNA virus.Genome Research, 19 (8): 1441-1449.
Monier, A., Welsh, R. M., Gentemann, C., Weinstock, G., Sodergren, E., Armbrust, E. V., Eisen, J. A., and Worden, A. Z., 2012. Phosphate transporters in marine phytoplankton and their viruses: Cross-domain commonalities in viral-host gene exchanges.Environmental Microbiology, 14 (1): 162-176.
Nagasaki, K., Shirai, Y., Tomaru, Y., Nishida, K., and Pietrokovski, S., 2005. Algal viruses with distinct intraspecies host specificities include identical intein elements.Applied and Environmental Microbiology, 71: 3599-3607.
Nissimov, J. I., Worthy, C. A., Rooks, P., Napier, J. A., Kimmance, S. A., Henn, M. R., Ogata, H., and Allen, M. J., 2011a. Draft genome sequence of the coccolithovirus EhV-84.Standards in Genomic Sciences, 5 (1): 1-11.
Nissimov, J. I., Worthy, C. A., Rooks, P., Napier, J. A., Kimmance, S. A., Henn, M. R., Ogata, H., and Allen, M. J., 2011b. Draft genome sequence of the coccolithovirusEmiliania huxleyivirus 203.Journal of Virology, 85: 13468-13469.
Nissimov, J. I., Worthy, C. A., Rooks, P., Napier, J. A., Kimmance, S. A., Henn, M. R., Ogata, H., and Allen, M. J., 2012a. Draft genome sequence of four coccolithoviruses:Emiliania huxleyivirus EhV-88, EhV-201, EhV-207, and EhV-208.Journal of Virology, 86 (5): 2896-2897.
Nissimov, J. I., Worthy, C. A., Rooks, P., Napier, J. A., Kimmance, S. A., Henn, M. R., Ogata, H., and Allen, M. J., 2012b. Draft genome sequence of the coccolithovirusEmiliania huxleyivirus 202.Journal of Virology, 86: 2380-2381.
Nissimov, J. I., Jones, M., Napier, J. A., Munn, C. B., Kimmance, S. A., and Allen, M. J., 2013. Functional inferences of environmental coccolithovirus biodiversity.Virologica Sinica, 28 (5): 291-302.
Nissimov, J. I., Napier, J. A., Kimmance, S. A., and Allen, M. J., 2014. Permanent draft genomes of four new coccolithoviruses:EhV-18, EhV-145, EhV-156 and EhV-164.Marine Genomics, 15: 7-8.
Pagarete, A., Allen, M. J., Wilson, W. H., Kimmance, S. A., and de Vargas, C., 2009. Host-virus shift of the sphingolipid pathway along anEmiliania huxleyibloom: Survival of the fattest.Environmental Microbiology, 11 (11): 2840-2848.
Pagarete, A., Lanzén, A., Puntervoll, P., Sandaa, R. A., Larsen, A., Larsen, J. B., Allen, M. J., and Bratbak, G., 2012. Genomic sequence and analysis of EhV-99B1, a new coccolithovirus from the Norwegian Fjords.Intervirology, 56:60-66.
Rickaby, R. E. M., Bard, E., Sonzogni, C., Rostek, F., Beaufort, L., Barker, S., Rees, G., and Schrag, D. P., 2007. Coccolith chemistry reveals secular variations in the global ocean car-bon cycle?Earth and Planet Science Letters, 253 (1-2): 83-95.
Ridgwell, A., and Zeebe, R. E., 2005. The role of the global carbonate cycle in the regulation and evolution of the Earth system.Earth and Planet Science Letters, 234 (3-4): 299-315.
Rohwer, F., and Thurber, R. V., 2009. Viruses manipulate the marine environment.Nature, 459 (14): 207-212.
Rowe, J. M., Fabre, M. F., Gobena, D., Wilson, W. H., and Wilhelm, S. W., 2011. Application of the major capsid protein as a marker of the phylogenetic diversity ofEmiliania huxleyi viruses.FEMS Microbiology Ecology, 76: 373-380.
Schroeder, D. C., Oke, J., Hall, M., Malin, G., and Wilson, W. H., 2003. Virus succession observed during anEmiliania huxleyiBloom.Applied and Environmental Microbiology, 69 (5):2484-2490.
Schroeder, D. C., Oke, J., Malin, G., and Wilson, W. H., 2002. Coccolithovirus (Phycodnaviridae): Characterisation of a new large dsDNA ) algal virus that infectsEmiliania huxleyi.Archives of Virology, 147: 1685-1698.
Sorensen, G., Baker, A. C., Hall, M. J., Munn, C. B., and Schroeder, D. C., 2009. Novel virus dynamics in anEmiliania huxleyibloom.Journal of Plankton Research, 31 (7): 787-791.
Sutterwala, S. S., Creswell, C. H., Sanyal, S., Menon, A. K., and Bangs, J. D., 2007. De novo sphingolipid synthesis is essential for viability, but not for transport of glycosylphosphatidylinositol-anchored proteins, in African trypanosomes.Eukaryotic Cell, 6 (3): 454-464.
Suttle, C. A., 2005. Viruses in the sea.Nature, 437: 356-361.
Takemura, M., 2002. Poxviruses and the origin of the eukaryotic nucleus.Journal of Molecular Evolution, 52 (5): 419-425.
Van Etten, J. L., 2011. Another really, really big virus.Viruses, 3 (1): 32-46.
Vardi, A., Haramaty, L., Van Mooy, B. A. S., Fredricks, H. F., Kimmance, S. A., Larsen, A., and Bidle, K. D., 2012. Host-virus dynamics and subcellular controls of cell fate in a natural coccolithophore population.Proceedings of the National Academy of Sciences, 109 (47): 19327-19332.
Vardi, A., Van Mooy, B. A. S., Fredricks, H. F., Popendorf, K. J., Ossolinski, J. E., Haramaty, L., and Bidle, K. D., 2009. Viral glycosphingolipids induce lytic infection and cell death in marine phytoplankton.Science, 326 (5954): 861-865.
Vogt, M., Steinke, M., Turner, S., Paulino, A., Meyerhfer, M., Riebesell, U., LeQuéré, C., and Liss, P., 2008. Dynamics of dimethylsulphoniopropionate and dimethylsulphide under different CO2concentrations during a mesocosm experiment.Biogeosciences Discuss, 5: 407-419.
Wilson, W. H., Schroeder, D. C., Allen, M. J., Holden, M. T. G., Parkhill, J., Barrell, B. G., Churcher, C., Hamlin, N., Mungall, K., Norbertczak, H., Quail, M. A., Price, C., Rabbinowitsch, E., Walker, D., Craigon, M., Roy, D., and Ghazal, P., 2005. Complete genome sequence and lyticphase transcription profile of a coccolithovirus.Science, 309: 1090-1092.
Wilson, W. H., Tarran, G. A., Schroeder, D., Cox, M., Oke, J., and Malin G., 2002. Isolation of viruses responsible for the demise of anEmiliania huxleyibloom in the English Channel.Journal of the Marine Biological Association of the United Kingdom, 82 (3): 369-377.
Wolfe, G. V., Steinke, M., and Kirst, G. O., 1997. Grazing activated chemical defense in a unicellular marine alga.Nature, 387: 894-897.
Zhang, Y. F., Liu, J. W., Zhang, Z. L., and Dong, S. L., 2010. Cloning and bioinformatic analysis of thioredoxin-like protein gene in marine coccolithophoridEmiliania huxleyivirus.Oceanologia et Limnologia Sinica, 40 (2): 294-297.
Zhaxybayeva, O., and Gogarten, J. P., 2007. Horizontal gene transfer, gene histories, and the root of the tree of life. In:Planetary Systems and the Origins of Life. Cambridge Uiversity Press, UK, 178-192.
(Edited by Ji Dechun)
(Received March 13, 2014; revised July 12, 2014; accepted August 17, 2014)
© Ocean University of China, Science Press and Springer-Verlag Berlin Heidelberg 2015
* Corresponding author. E-mail: microzh@xmu.edu.cn
 Journal of Ocean University of China2015年2期
Journal of Ocean University of China2015年2期
- Journal of Ocean University of China的其它文章
- Effect of Dietary Lipid on the Growth,Fatty Acid Composition and Δ5 Fads Expression of Abalone(Haliotis discus hannai Ino)Hepatopancreas
- Species Composition and Diversity of Macrobenthos in the Intertidal Zone of Xiangshan Bay, China
- Evaluation of Cytotoxicity and Genotoxicity of Insecticide Carbaryl to Flounder Gill Cells and Its Teratogenicity to Zebrafish Embryos
- Purification of a Diatom and Its Identification to Cylindrotheca closterium
- Mechanical Stress Induces Neuroendocrine and Immune Responses of Sea Cucumber (Apostichopus japonicus)
- Preparation of κ-carra-Oligosaccharides with Microwave Assisted Acid Hydrolysis Method
