Effects of Transgenic DREB Toybean Dongnong50 on Diversity of Soil Nitrogen-fixing Bacteria
Zhang Chun-miao,Dong Lei,Jin Yu,and Qu Juan-juan
College of Resources and Environment,Northeast Agricultural University,Harbin 150030,China
Introduction
China was once the world's foremost soybean producing country with 71.5% of the world total output,which is far more than the sum of the other countries.Now China become a net soybean importer due to the price advantage of genetically modified soybeans for processing enterprises.Drought is a bottleneck preventing soybean production in Northeast of China.Dehydration responsive element binding (DREB) is a kind of transcription factor that regulates the expression of stress tolerance-related genes in response to drought,high salinity and cold stress in plants (Kumar and Rosen,2006).The yield of DREB transgenic soybean can be greatly increased due to its tolerance to drought.Concerns have been raised because the release of transgenic plants worldwide would endanger ecologic system and their replacement of traditional crops would reduce biodiversity.
The response of soil biodiversity to transgenic plant has been investigated in many studies.Engineered gene or its products may be released into soil via plant litter,sloughed and root exudation.Horizontal gene transfer to indigenous microorganism in soil will then change the community structure and diversity of soil microbes (Gebhard and Smalla,1998).In addition,released gene products by transgenic plants were hard to degrade and would persist for a long time (Kremer and Means,2005).Effects on soil microorganisms may arise from changes of physiological and biochemical characteristics of transgenic plants and variation of the transgenic genes (Melnitchouck and Leinweber,2006).However,minor alterations in the diversity of the microbial community could affect the health and function of soil ecosystem (Angle,1994).In recent years,many researches about the effects of genetically modified plants (GMPs) on soil microorganisms have been executed.Donegan et al.(1999) found that the metabolic fingerprints of the microbial community of transgenic alfalfa were different from those of non-transgenic lines.Cowgill et al.(2002) discovered that transgenic potato not only restrained the growth of soil bacteria and fungi but also reduced their abundance.According to Means and Kremer (2007),glyphosate-resistant (GR) soybean increased microbial activity and the growth of rootassociated Fusarium spp.On the contrary,many studies revealed transgenic plants had no deleterious effects on soil microorganisms transgenic,such as Bt and herbicide-resistant transgenic maize (Devare and Londoño-R,2007;Schmalenberger and Tebbe,2002).Moreover,the research of Liu et al.(2008) revealed that no significant effect was found on key microbial processes or microbial community composition in rice rhizosphere over two years.
Nitrogen availability can affect the rate of key ecosystem process including primary production and decomposition (Vitousek and Howarth,1991;Tilman,1988).Nitrogen-fixing microorganisms can transform molecular nitrogen in the atmosphere into inorganic nitrogen used by plants.nifH gene encoding the iron protein subunit of nitrogenase,which was highly conservative among nitrogen-fixing bacteria,is appropriate to be used to research the diversity of nitrogen-fixation bacterial community (Jerome et al.,2002).Zhao (2006) researched the influence of transgenic soybean on soil nitrogen transformation and relevant bacterial diversity,and found that the transgenic soybean reduced the biodiversity index of soil at the vigorous stage.Research on the effects of Round Ready soybean (RRS) on ammonia-oxidizing bacteria (AOB) communities revealed that Shannon-Wiener diversity indexes (Dsh) and evenness indexes (Jsh) related to AOB in RRS rhizospheric soils were lower than those of near-isogenic counterparts (RRS-S) (Xu et al.,2008).However,Lai et al.(2010) explored the effects of transgenic soybean on the nitrogenfixing bacteria in rhizospheric soil,found that it had no adverse effects on the diversity of nitrogenfixing bacteria in rhizospheric soils.Zhou et al.(2010) reported that transgenic soybean with two fungus-resistant genes had no significant effects on the amounts and community structure of microbes in its rhizospheric soils.GMPs may have different ecological effects due to the characters of the transferred gene,integration sites and copy numbers (Yuan et al.,2005).Because of the complexity of the soil ecosystem structure and microbial diversity,we should assess the safety and reliability of the genetically modified plants deeply.The present work aimed to research the effects of transgenic soybean on diversity of nitrogen-fixing bacteria using PCR-DGGE technology.The results would provide references for risk assessment of transgenic soybean.
Materials and Methods
Soil sample
Soil was collected from Horticultural Experiment Station of Northeast Agricultural University.Physicochemical properties of initial soil were determined with routine methods recommended by SSSA Soil Science Society of America and listed in Table 1.Seeds of transgenic DREB soybean Dongnong50 and its near-isogenic nontransgenic counterparts were provided by Key Lab of Soybean Biology of Education Ministry.

Table1 Main physicochemical properties of initial soil
Pot experiments
Pot experiments were conducted in Horticultural Experiment Station of Northeast Agricultural University.Soybeans were sowed in May 2012 and harvested in September 2012.There were four treatments: A-transgenic soybean under normal water condition;B-non-transgenic soybean under normal water condition;C-transgenic soybean under drought stress condition;D-non-transgenic soybean under drought stress condition.Each treatment had 20 pots.Five seeds were sowed in each pot and thinned to one seedling after emergence.Each pot of group A and B was irrigated 1 000 mL of water every two to three days.The group of C and D were under drought stress condition,whose water content was 50%-55% of normal water condition measured by Time-domain reflectometry.
Soil samples of each treatment were collected randomly from the rhizosphere (in the top 5 cm) of five pots at pre-sowing stage,seeding stage,flowering stage,podding stage,seed filling stage,and harvest time.All the samples of one treatment were mixed thoroughly into one representative sample which was then sieved (4 mm mesh) and stored at 4℃.
Soil total DNA extraction
Soil DNA was extracted from 0.5 g fresh soil with modified method described according to Zhou et al.(2010).DNA extraction was further purified using DNA purification kit (TIANGEN DNA gel extraction kit) to remove the humic substance according to the instruction manual.All purified products were analyzed on 1% agarose gel electrophoresis.Then soil samples were stored at –80℃ until use.
PCR amplification
Nested PCR was used to amplify target nifH sequence;the primers and reaction conditions are shown in Table 2.The reaction mixture of first-round PCR reaction consisted of 10 ng DNA template,2 μL 10 mmol • L-1primers,5 μL 10×buffer,0.5 μL Taq DNA polymersase (TaKaRa) in a total volume of 50 μL.The reaction mixture of second-round PCR reaction consisted of 1 μL of the first-round PCR product as template,2 μL of 10 mmol • L-1primers,5 μL of 10×buffer,0.5 μL of Taq DNA polymersase (TaKaRa) in a total volume of 50 μL.
DGGE analysis
DGGE was performed with D Code Universal Mutation System (Bio-Rad).25 μL of PCR products were loaded into 8% (w/v) polyacrylamide (37.5:1,acrylamide: bisacrylamide) gels with a denaturing gradient of 40%-65%.
The gel was run in 1× Tris-acetate EDTA buffer at a constant voltage of 70 V for 16 h at 60℃.After the electrophoresis,the gel was washed twice with distilled water and then immediately fixed with fixation solution (10% ethanol+0.5% acetic acid) for 15 min.Followed by incubation in silver nitrate solution (0.2% AgNO3+10% ethanol+0.5% acetic acid) avoiding light for 3 min.The gel image was captured with the Gel Doc 2000 System of Bio-Rad.

Table2 Sequence of primers and PCR conditions
NifH gene sequencing and phylogenetic analysis
Selected predominant bands were excised from the DGGE gel and transferred into 1.5 mL sterile tubes containing 20 μL of double distilled water after washed twice.The tubes were stored overnight at 4℃.4 μL of the solution were used as template in PCR using the former protocol.The resulting PCR products were rerun on a gel to demonstrate single band that migrated to the same position,thus avoiding other DGGE bands had also been inadvertently excised.Purified bands were excised for a second time,and then were eluted and reamplified with the primers of PolF and AQER.PCR products were analyzed by 1% agarose gel electrophoresis,and then the products were sequenced by Shanghai Sangon Biological Engineering Technology Services Co.,Ltd.
Data and DGGE profile analyses
SPSS18.0 was used for all data analysis.Quantity One software 4.0 (Bio-Rad) was used to analyze the DGGE profile.All the sequences were evaluated based on BLAST alignments (Altschul et al.,1997) and a phylogenetic tree was constructed with MEGA 5.0.
Results
Soil total DNA extraction
High quality DNA template was the basis for molecular ecology study of microbial community structure.The size of total DNA fragment was 23.1 kb which was shown in Fig.1.

Fig.1 Agarose gel electrophoresis of soil total DNA
PCR amplification of nifH gene
The first-round PCR products of nifH gene were almost invisible,but the second-round PCR products were distinct and the size was about 360 bp(Fig.2).

Fig.2 PCR products of nifH gene with soil total DNA as template
DGGE profile analysis
DGGE profile of nifH gene of nitrogen-fixing bacteria revealed that band g9 was shared by all samples.g16 and g20 were specific bands shared by sample A2.g4,g11 and g15 were specific bands shared by Sample B2.g6 was specific band shared by Sample D3.The emergence of specific bands had closely related to temperature,soil water content and pH.The band number varied with growing period,and higher diversity was shown in DGGE profiles of the nitrogen-fixing bacteria at podding stage and seed filling stage.
Shannon-Wiener index analysis
Shannon-Wiener index of DGGE profile for different treatments is presented in Table 3,which showed that diversity of microbial community varied with the growth stage in all treatments but no significance was found between the treatments of transgenic soybean and non-transgenic soybean under normal water condition and drought stress condition.It indicated that planting transgenic soybean had no notable effects on the diversity of soil nitrogen-fixing bacteria in the whole growth period compared with non-transgenic soybean.Saxena and Florest (2002) also revealed that the cultivation of Bt maize did not produce a significant impact on major microflora of bacteria,actinomycetes and fungi in rhizosphere soil.
Similarity analysis
Lane 6 was as the standard lane compared with the other lanes in DGGE patterns (Fig.4).There were low diversity between Lane 5 and Lane 6,Lane 7 and Lane 8,Lane 9 and Lane 10 compared with the standard lane.There were high diversity in other lanes from the soil samples of the transgenic soybean and non-transgenic soybean under normal water condition and drought stress condition at the same period.

Table3 Shannon-Wiener index of DGGE profile
The similarity of Lane 1 and Lane 2,Lane 5 and Lane 6,Lane 9 and Lane 10,Lane 13 and Lane 14,Lane 17 and Lane 18 was 72.7%,53.3%,35.2%,78.3% and 76.3%,respectively (Fig.5).The band numbers and the diversity of Land 5 and Lane 9 were respectively less than Lane 6 and Lane 10.The results showed that transgenic soybean under normal water condition decreased the diversity of the soil nitrogen-fixing bacteria at the seeding stage and flowering stage,and there was no significant difference between diversity of the transgenic soybean and nontransgenic soybean under normal water and drought stress condition at the same period.In addition,Figs.3-5 also displayed the similarity of Lane 3 and Lane 4,Lane 7 and Lane 8,Lane 11 and Lane 12,Lane 15 and Lane 16,Lane 19 and Lane 20 which was 80.1%,39%,75.2%,60% and 69.8%,respectively.The band number and the diversity of Land 7 was less than that of Lane 8.The above results illustrated that the diversity of the soil nitrogen-fixing bacteria was affected by the transgenic soybean under drought stress condition at seeding stage,but no effects on other stages.The divergence may be caused by the difference of soybean root exudates including amounts of amino acids and carbohydrates which have an impact on rhizosphere microorganisms (Kremer and Means,2005).

Fig.3 DGGE profiles of soil nitrogen-fixing bacteria in different treatments

Fig.4 Comparison of DGGE patterns

Fig.5 Jaccard similarity matrix of soil nitrogen-fixing bacteria of different samples
Cluster analysis
On the basis of the migration positions and the band intensity of different treatments in DGGE fingerprinting,cluster analysis of community structure of soil nitrogen-fixing bacteria from different samples was performed with UPGAMA method.In Figs.3-6,the similarities of Lanes 5 and 6,Lanes 7 and 8,Lanes 9 and 10 were lower than 60%.The analysis results demonstrated that transgenic soybean and non-transgenic soybean under normal water condition had low similarity in community structure of soil nitrogenfixing bacteria at the seeding stage and flowering stage.Furthermore,transgenic soybean and nontransgenci soybean under drought stress condition had low similarity at the seeding stage.Soybean is a water requirement crop whose normal growth should be under 70% field moisture capacity.In flowering stage and podding stage,water shortage will adversely affect soybean growth,resulting in flower and pod abscission.Moreover,soil bacterial diversity was corresponding changed with soybean growth status (Wang et al.,2013).
nifH gene sequencing and phylogenetic analysis
Rhizosphere associative dinitrogen fixation could be a valuable source of nitrogen in many nitrogen limited natural ecosystems.nifH is a dinitrogenase reductase encoding gene that is very conservative in nitrogen-fixing organisms within the symbiotic community (Powell et al.,2009).In Fig.3,21 prominent bands of nifH PCR-DGGE product were retrieved from DGGE gel,and subsequently reamplified and sequenced.
All sequences had been deposited in GenBank database with accession numbers from JQ437983 to JQ438002 and JN851858.The most similar se-quences in GenBank database were determined by BLASTN search tool (Altschul et al.,1997) and shown in Table 4.The results suggested that all the sequences were very similar to the sequence of nifH gene.

Fig.6 Cluster analysis (UPGMA,Dice coefficient of similarity) generated by DGGE profile

Table4 GenBank accession number and identity of sequences from DGGE

Continue
A NJ tree based on nifH gene sequences was constructed using MEGA software (Fig.7).A total of 17 bacterial sequences were derived from DGGE bands,which were excised from the full-gradient range of the gel,and sequenced.All sequences had been deposited in the GenBank database with accession numbers JQ438016 to JQ438029.The closest relative strains available in the GenBank database were obtained using Blastn Program.Phylogenic analysis revealed that g7,g13,g15 and g19 had a close relationship with Alphaproteobacteria,g12 had a close relationship with Azonexus,others were related to Betaproteobacteria and Burkholderia.
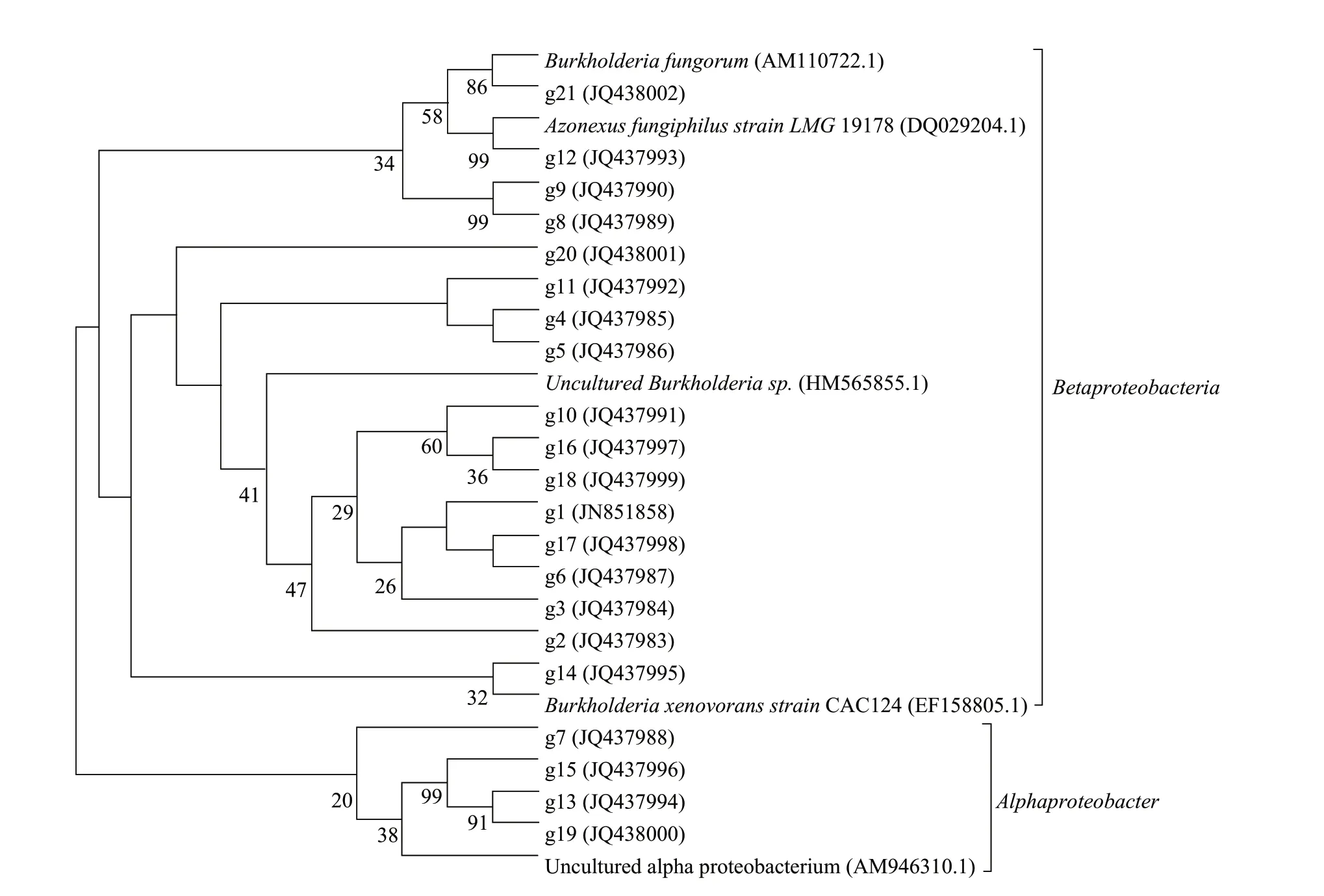
Fig.7 Phylogenetic tree based on nifH gene sequence
The excretions in rhizosphere of transgenic soybean may be different from that of non-transgenic soybean,inhibiting the growth of partial soil nitrogen-fixing bacteria,while stimulating others'growth (Yuan et al.,2005).Transgenic soybean did not make much impact on the community structure under normal water condition in podding stage and seed filling stage,harvest time,while also had no any effect on the community structure under drought stress condition in flowering stage,podding stage and seed filling stage,harvest time.Changes in the composition of excretions in rhizosphere of transgenic soybean may been influenced by difference of environmental factors of moisture,temperature,nutrients,and pH at different growth stage (Finlay and Maberly,1997;Groffman and Bohlen,1999).
Conclusions
This research explored the effects of transgenic soybean on the diversity of soil microbe especially nitrogen-fixing bacteria.In the present study,however,the soybean varieties did not significantly affect densities of nitrogen-fixing bacteria.The existence of the plant DREB transcription factors involved in stress response to the external environment would improve drought resistant of plants to maintain the photosynthetic intensity,extend the photosynthetic time and increase production (Xu et al.,2005;Liang,2004;Qiu,2009).The analysis of DGGE profile showed that no great difference was found in Shannon-Wiener index between the transgenic soybean and non-transgenic soybean under normal water condition and drought stress condition in the whole growth period.But significant variability in community structure was observed between the transgenic soybean and non-transgenic soybean under normal water condition in seeding stage and flowering stage.There were less nitrogen-fixing bacteria of transgenic soybean than non-transgenic soybean under normal water condition in seeding and flowering stage.Moreover,they both had their own advantage species.g16 and g20 were shared by the samples of transgenic soybean under normal water condition in seeding stage.g3,g4,g5,g11,and g15 were shared by soil samples of nontransgenic soybean under normal water condition in flowering stage.There was a distinct difference in community structure between transgenic soybean and non-transgenic soybean under drought stress condition in seeding stage,but no differences on other stages.
Agarwal P K,Agarwal P,Reddy M K,et al.2006.Role of DREB transcription factors in abiotic and biotic stress tolerance in plants.Plant Cell Reports,25(12): 1263-1274.
Altschul S F,Madden T L,Schaffer A A,et al.1997.Gapped BLAST and PSI-BLAST: a new generation of protein database search programs.Nucl Acids Res,25(17): 3389-3402.
Angle J S.1994.Release of transgenic plants: biodiversity and population-level considerations.Molecular Ecology,3: 45-50.
Cowgill S E,Bardgett R D,Kiezebrink D T,et al.2002.The effect of transgenic nematode resistance on non-target organisms in the potato rhizosphere.Journal of Applied Ecology,39: 915-923.
Devare M,Londoño-R L M,Thies J E.2007.Neither transgenic B tmaize (MON863) nor tefluthrin insecticide adversely affect soil microbial activity or biomass: a 3-year field analysis.Soil Biology and Biochemistry,39(8): 2038-2047.
Donegan K K,Seidler R J,Doyle J D,et al.1999.A field study with genetically engineered alfalfa inoculated with recombinant sinorhizobium meliloti: effects on the soil ecosystem.Journal of Applied Ecology,36: 920-936.
Dubouzet J G,Sakuma Y,Ito Y,et al.2003.DREB genes in rice,Oryza sativa L.,encode transcription activators that functioning drought,high-salt and cold-responsive gene expression.Plant Journal,33(4): 751-763.
Finlay B J,Maberly S C,Cooper J I.1997.Microbial diversity and ecological function.Oikos,80: 209-213.
Gebhard F,Smalla K.1998.Transformation of Acinetobacter sp.strain BD413 by transgenic sugar beet DNA.Applied and Environmental Microbiology,64(4): 1550-1555.
Groffman P M,Bohlen P J.1999.Soil and sediment biodiversity cross system comparisons and largescale effects.Bioscience,49(2): 139-148.
Hao X Y,Chen M,Xu H J,et al.2005.GH-DREB gene transformation of wheat drought physiological indicators identified and progeny transgenic plants.Southwest Agricultural Sciences,18(5): 616-620.
Jerome H N F,Sonia T,Sylvie T C,et al.2002.NifH gene diversity in the bacterial community associated with the rhizosphere of Molinia coerulea,an oligonitrophilic perennial grass.Environmental Microbiology,4(2): 477-481.
Kremer R J,Means N E.2005.Glyphosate affects soybean root exudation and rhizosphere microorganisms.International Journal of Environmental Analytical Chemistry,85: 1165-1174.
Kumar K,Rosen C J,Ruselle M P.2006.Enhanced protease inhibitor expression in plant residues slows nitrogen mineralization.Agronomy Journal,98: 514-521.
Lai X,Zhang Y S,Zhao S,et al.2010.Effects of transgenic soybean on the diversity of nitrogen fixing-bacteria in rhizosphere soil.Chinese Journal of Ecology,29(9): 1736-1742.
Liang X X.2004.Study on improvement of salt tolerance by transformation with transcription factor DREB in wheat.Academy of Agricultural Sciences,Beijing.
Liu L,Zhu K,Yang Y,et al.2008.Molecular cloning,expression profiling and trans-activation property studies of a DREB 2-like gene from chrysanthemum.Journal of Plant Research,12(1): 215-226.
Liu W,Hao L H,Wu W,et al.2008.Transgenic Bt rice does not affect enzyme activities and microbial composition in the rhizosphere during crop development.Soil Biology and Biochemistry,40(3): 475-486.
Means N E,Kremer R J,Ramsier C.2007.Effects of glyphosate and foliar amendments on activity of microorganisms in the soybean rhizosphere.Journal of Environmental Science and Health.Part.B,Pesticides,Food Contaminants,and Agricultural Wastes,42(2): 125-132.
Melnitchouck A,Leinweber P,Eckhardt I.2006.Pyrolysis-field ionization mass spectrometry of genetically modified plants on soilorganisms.Environmental Biosafety Research,5(1): 37-46.
Nap J-P,Metz P L J,Escaler M,et al.2003.The release of genetically modified crops into the environment: an overview of current status and regulations.Plant J,33(7): 1-18.
Ni Z Y,Xu Z S,Li L C,et al.2008.Mechanism and application prospect of DREB transcription factors in plant stress resistance.Journal of Triticeae Crops,28(2): 1100-1106.
Powell J R,Levy-Booth D J,Gulden R H,et al.2009.Effects of enetically modified,herbicide-tolerant crops and their management on soil food web properties and crop litter decomposition.J Appl Ecol,46: 388-396
Quiviger B,Franche C,Lutfalla G,et al.1982.Cloning of nitrogen fixation (nif) gene cluster of Azospirillum brasilense.Biochimie,64(7): 495-502.
Qiu M.2009.Different strains of wheat DREB gene transfer physiological and biochemical characteristics of drought.Shandong Agricultural University,Jinan.
Saxena D,Florest S,Stotzky G.2002.Bt toxin is released in root exudates from 12 transgenic corn hybrids representing three trans forma tionevents.Soil Biology and Biochemistry,3(4): 133-137.
Schmalenberger A,Tebbe C C.2002.Bacterial community composition in the rhizosphere of a transgenic,herbicide-resistant maize (Zea mays) and comparison to its non-transgenic cultivar Bosphore.FEMS Microbiology Ecology,40(1): 29-37.
Tilman D.1988.Plant strategies and dynamics and structure of plant communities.Princeton University Press,Princeton N J.pp.26-28.
Vitousek P M,Howarth RW.1991.Nitrogen limitation on land and in the sea: how can it occur? Biogeochemistry,13(3): 87-115.
Wang L J,Li G,Zhao J N,et al.2013.Effects of transgenic soybean rhizosphere soil microbial community functional diversity.Journal of Agro-Environment Science,32(2): 290-298
Watve M G,Tickoo R,Jog M M,et al.2001.How many antibiotics are produced by the genus streptomyces.Archives of Microbiology,17(6): 386-390.
Xu G H.2010.Effects of RRS on the diversity of AOB and AOA in rhizospheric soils.Northeast Agricultural University,Harbin.
Xu S,Hu J,Chen Y H,et al.2008.Progress DREB transcription factor.Journal of Agricultural Biotechnology,16(4): 706-713.
Yang H J,Xiao Q M,Liu A Y.2005.Soil microbial diversity and its action.Journal of Nanhua University (Science and Technology),12(4): 21-31.
Yuan H X,Zhang J Z,Guo J F,et al.2005.Activities of microbes and enzymes in soil after growing transgenic rice with two extra antifungus genes.Acta Pedologoca Sinica,42(1): 122-126.
Zhao G.2006.Study on soil nitrogen transformed and relevant bacterium's diversity influence of transgenic soybean.Northeast Agricultural University,Harbin.
Zhou L,Shu C L,Hung W K,et al.2010.Community structure of microbes in rhizosphere soil of transgenic soybean carrying two fungus-resistant genes.Chinese Journal of Applied Environmental Biology,16(4): 509-514.
 Journal of Northeast Agricultural University(English Edition)2015年1期
Journal of Northeast Agricultural University(English Edition)2015年1期
- Journal of Northeast Agricultural University(English Edition)的其它文章
- SWOT Analysis and Countermeasures of Ecological Agricultural Development of Jianshan Farm
- Evolution Analysis About Soybean MIR166 Family
- Influence of FSH Treatment on Expression of CDC25A,TSSK3 and P53 in Vitro Cultured Sertoli Cells of Calf
- Dynamic Analysis of Nitric Oxide and Total Oxidant Capacity in Cow Uterine Secretion with Subclinical Endometritis
- Antioxidant Activities of Nine Selected Culinary Spices from China
- Fault Line Selection Method Considering Grounding Fault Angle for Distribution Network
