产芳香腈水解酶的恶臭假单胞菌Pseudomonas putida CGMCC3830的筛选、鉴定及发酵优化
朱小燕,龚劲松,李恒,陆震鸣,周哲敏,史劲松,许正宏
产芳香腈水解酶的恶臭假单胞菌Pseudomonas putida CGMCC3830的筛选、鉴定及发酵优化
朱小燕1,2,龚劲松1,2,李恒1,陆震鸣1,周哲敏2,史劲松1,许正宏1,2
1 江南大学药学院,江苏 无锡 214122
2 江南大学 工业生物技术教育部重点实验室,江苏 无锡 214122
近年来微生物腈水解酶水解腈类化合物制备有机酸已逐步受到关注。本研究分离到一株表现出较高腈水解酶活力的细菌菌株,通过形态学、生理生化实验以及16S rRNA基因序列分析将其鉴定为恶臭假单胞菌Pseudomonas putida CGMCC3830。结合单因素及响应面法对该菌株产腈水解酶的发酵条件进行了优化,获得最适培养条件为:甘油13.54 g/L,胰蛋白胨11.59 g/L,酵母粉5.21 g/L,KH2PO41 g/L,NaCl 1 g/L,脲1 g/L,初始pH 6.0及培养温度30 ℃。通过优化,酶活由2.02 U/mL提升至36.12 U/mL。对该菌株底物特异性的考察结果表明,恶臭假单胞菌腈水解酶对芳香族腈类化合物具有较高的水解活力。将其应用于烟酸的生物合成中,2 mg/mL游离细胞能90 min内将20.8 g/L 3-氰基吡啶彻底转化,制备得到相应烟酸。这些结果表明恶臭假单胞菌P. putida CGMCC3830在烟酸的规模化生产中具有一定的应用潜力。
腈水解酶,恶臭假单胞菌,3-氰基吡啶,烟酸,发酵优化
Biocatalysis has long been considered to be an attractive choice in chemical synthesis compared with traditional metallo- and organo-catalysis due to its specificity, and “green” properties[1-3]. As one of the most valuable biocatalysts, nitrilase mediates the nitriles hydrolysis for production of corresponding carboxylic acids, which could be used as precursors for pharmaceuticals, fine chemicals, and food additives[4-5]. It has received considerable industrial interests owing to the mild reaction conditions, as well as excellent catalytic efficiency and selectivities[6-7]. Consequently, an increasing number of academic institutions and companies (Lonza, Dow, Diversa, BASF, and DSM) are exploring the synthetic potential of nitrilases for bioproduction of carboxylic acids with commercial value, such as nicotinic acid, isonicotinic acid, (R)-(-)-mandelic acid, p-hydroxybenzoic acid, acrylic acid, and some amino acids[8-12].
The applications of nitrilase as a general biocatalyst are largely unexploited, as compared to the successful utilization of lipase and esterase[13]. One of the bottlenecks in development of such a specific biotransformation is to find an appropriate biocatalyst. Bacteria, fungi, and plants are excellent sources of nitrilase, with bacterial species as the main source[14]. Furthermore, the genus Pseudomonas was found to be a promising source for nitrilases. The first bacterial nitrilase was discovered in Pseudomonas sp., which could hydrolyze the ricinine nitrile group[15]. From then on, several bacteria from the genera Pseudomonas werereported to display nitrilase activity. The nitrilases from P. fluorescens DSM 7155[16], P. fluorescens EBC 191[17]and P. putida MTCC 5110[18]showed preference for arylacetonitriles, such as phenylacetonitrile and mandelonitrile. Aliphatic nitriles, such as fumarodinitrile and succinonitrile, were proven to be efficient substrates for P. fluorescens Pf-5 nitrilase[19]. However, the nitrilases of these Pseudomonas strains exhibit minimal activity toward aromatic nitriles, such as 3-cyanopyridine and benzonitrile. To sum up, Pseudomonas strains with aromatic nitrilase activity have never been reported.
Since the potential of Pseudomonas nitrilase for nitriles biotransformation being clearly demonstrated, it is a necessary work to promote the nitrilase production from Pseudomonas strains through culture conditions optimization. To date, studies on culture conditions for enhancing nitrilase production by microorganisms have been scanty. For optimization of culture conditions, the one-factor-ata-time (OFAT) method could be easily implemented and could be used for the selection of the significant parameters affecting enzyme yield[20]. Response surface methodology (RSM) can be utilized to investigate the interactions among factors and to obtain the optimal level of test variables[21-22].
The aim of this study was to screen a novel strain harboring nitrilase acitivity for conversion of 3-cyanopyridine into nicotinic acid. The isolated strain from environmental sources was identified as P. putida CGMCC3830. Subsequently, the nitrilase production was enhanced by optimization of culture conditons with a combination of OFAT and RSM, and the application performance of this strain was investigated and discussed. The strain was proven to be a potential biocatalyst for nitrile hydrolysis and thus pave the way for establishment of an economical bioprocess, which could effectively convert 3-cyanopyridine into nicotinic acid in a commercial scale.
1 Materials and methods
1.1 Screening of microorganisms
Soil samples were collected from Jiangsu Province, China. Approximately 1 g of each soil sample was added to 50 mL of a nitrogen-free mineral medium (per liter: glucose 5 g, KH2PO41 g, MgSO40.1 g, FeSO4·7H2O 0.02 g, CaCl20.02 g, NaCl 1 g, ε-caprolactam 1 g, and pH 7.0) supplemented with 0.5 g/L 3-cyanopyridine as sole nitrogen source and incubated at 30 ℃ for 3 d. After enrichment cultivation, the suspension was diluted and spread on agar plates to isolate pure colonies. The isolated colonies were further incubated in the fermentation medium (per liter: glucose 10 g, peptone 5 g, yeast extract 5 g, NaCl 1 g, KH2PO42 g, MgSO40.1 g, FeSO4·7H2O 0.03 g, ε-caprolactam 1 g, and pH 7.0) at 30 ℃ with shaking at 120 r/min for 48 h for enzyme production. The resulting cells were harvested by centrifugation (10 000 × g, 10 min) and then washed twice with 100 mmol/L potassium phosphate buffer (pH 7.2). The washed cells were resuspended in the same buffer for further study.
1.2 Taxonomic identification
The isolate was identified based on its morphology, biochemical and physiological characteristics, as well as 16S rRNA gene sequence. Morphological properties were observed under an optical microscope and a scanning transmission electron microscope (SEM). Biochemical and physiological characterization assays were conducted as described in Bergey’s Manual of Systematic Bacteriology[23]. The 16S rRNA gene sequence of the isolate was amplified via polymerase chain reaction by using two universal primers, P0(5'-GAGAGTTTGATCCTGGCTCAG-3') and P6(5'-CTACGGCTACCTTGTTACGA-3'). A similarity search for the 16S rRNA gene sequence was conducted using the Basic Local Alignment Search Tool provided by the National Center for Biotechnology Information. A total of 16S rRNAgene sequence and related sequences obtained from the GenBank database were aligned using CLUSTAL X version 1.8[24]. A phylogenetic tree was constructed based on the homologous 16S rRNA gene sequence by using the Molecular Evolutionary Genetics Analysis software version 5.02.
1.3 Nitrilase activity assay
Nitrilase activity was assayed with 3-cyanopyridine as substrate using P. putida CGMCC3830 as whole-cell biocatalyst. The standard assay reaction was conducted in 2.0 mL of potassium phosphate buffer (100 mmol/L, pH 7.2) containing 50 mmol/L 3-cyanopyridine and 1 mg resting cells (dry cell weight) at 30 ℃ for 10 min. The reaction was terminated by adding 0.2 mL of 2 mol/L HCl, after which the reaction mixture was centrifuged. The 3-cyanopyridine and nicotinic acid in the supernatant were analyzed by high-performance liquid chromatography (HPLC). The ammonia in the supernatant was assayed using an ultraviolet spectrophotometer (Mapada UV-1800, Shanghai, China) according to phenol-hypochlorite reaction[25].
One unit of the enzyme activity was defined as the amount of enzyme that catalyzed the formation of nicotinic acid or ammonia at the rate of 1 μmol per min under the standard assay conditions. Specific activity was expressed as units per mg of dry cell. Total activity was expressed as units per mL of culture broth.
1.4 Optimization of culture conditions
Optimizations of various parameters for nitrilase production of P. putida CGMCC3830 were carried out using different medium components and cultivation conditions. The effects of different medium components on biomass and enzyme activity were evaluated by addition of various carbon sources, nitrogen sources, metal ions, inducers, and these components at different concentrations. In addition, the optimum cultivation conditions were investigated by varying the initial medium pH and cultivation temperature. The interactive influence of the significant factors was further investigated by RSM. The experiment was set up based on a central composite design (CCD). Design-Expert V8.0.6 software was utilized for the regression and graphical analysis of the obtained data.
1.5 Analytical methods
The growth of P. putida CGMCC3830 was estimated turbidimetrically, and the biomass (dry cell weight) was calculated from the predetermined standard curve relating the OD600to mg/mL (1 OD600= 0.40 mg/mL).
HPLC (Ultimate 3000, Dionex 3000, USA) was performed with a chromeleon system equipped with an Atlantis dC18 column (5.0 μm, 150 mm × 4.6 mm; Waters, USA) at a wavelength of 268 nm, column temperature of 30 ℃. The mobile phase was acetonitrile: 0.025% phosphoric acid (6/4, V/V) at a flow rate of 0.5 mL/min.
2 Results and discussion
2.1 Screening and identification of the nitrile-converting microorganism
Nitrilase has emerged as an efficient biocatalyst in green chemistry[26]. Novel and potential nitrilases from different sources have sprung up in recent years[11]. Utilizing 3-cyanopyridine as the sole nitrogen source, this work isolated several microbial strains and selected the bacterium with the highest activity towards 3-cyanopyridine.
The colonies of the isolated bacterium on a lysogeny broth plate were milky, yellow, convex, smooth, wet and fluorescent on King's B agar medium. The bacterium was gram negative with width ranging from 0.8 µm to 1 μm and length ranging from 1.5 µm to 2 μm. The flagellum was observed under SEM. Catalase and arginine dihydrolase reactions were positive, whereas starch hydrolysis, nitrate reduction, and liquefaction of gelatin were negative. The 16S rRNA gene sequenceof the bacterium was determined and then deposited in the GenBank database with accession number JQ086574. A phylogenetic tree was constructed based on the 16S rRNA gene sequence (Fig. 1). This strain was located in the same clade with P. putida (EF100617). Based on morphology, physiological and biochemical characteristics, as well as 16S rRNA gene sequence, the bacterium was identified as P. putida. The strain was deposited in the China General Microbiological Culture Collection Center with accession number CGMCC 3830; thus, it was designated as P. putida CGMCC3830.
2.2 Effects of carbon sources on nitrilase production
Effects of various carbon sources (10 g/L) were investigated for enhancing nitrilase production. Fig. 2 showed that sucrose, lactose, sorbitol, mannitol and soluble starch inhibited nitrilase formation, resulting in a specific activity of less than 0.61 U/mg. However, these carbon sources, particularly soluble starch, favored nitrilase formation of P. putida MTCC 5110[27]. Glycerol addition have a positive influence on cell growth (biomass 2.85 mg/mL) and nitrilase formation (specific activity 1.06 U/mg), which lead to the highest total activity of 3.02 U/mL, followed by D-fructose and glucose. The catabolite repression when using glycerol, D-fructose and glucose as carbon sources for P. putida MTCC 5110 nitrilase production[27]was not observed in this study. The optimal glycerol concentration was further investigated. The maximum nitrilase production was achieved with 10 g/L of glycerol. No improvement was observed with more than 10 g/L glycerol, whereas a negative effect was identified with 40 g/L glycerol. Finally, 10 g/L glycerol was selected for further study.
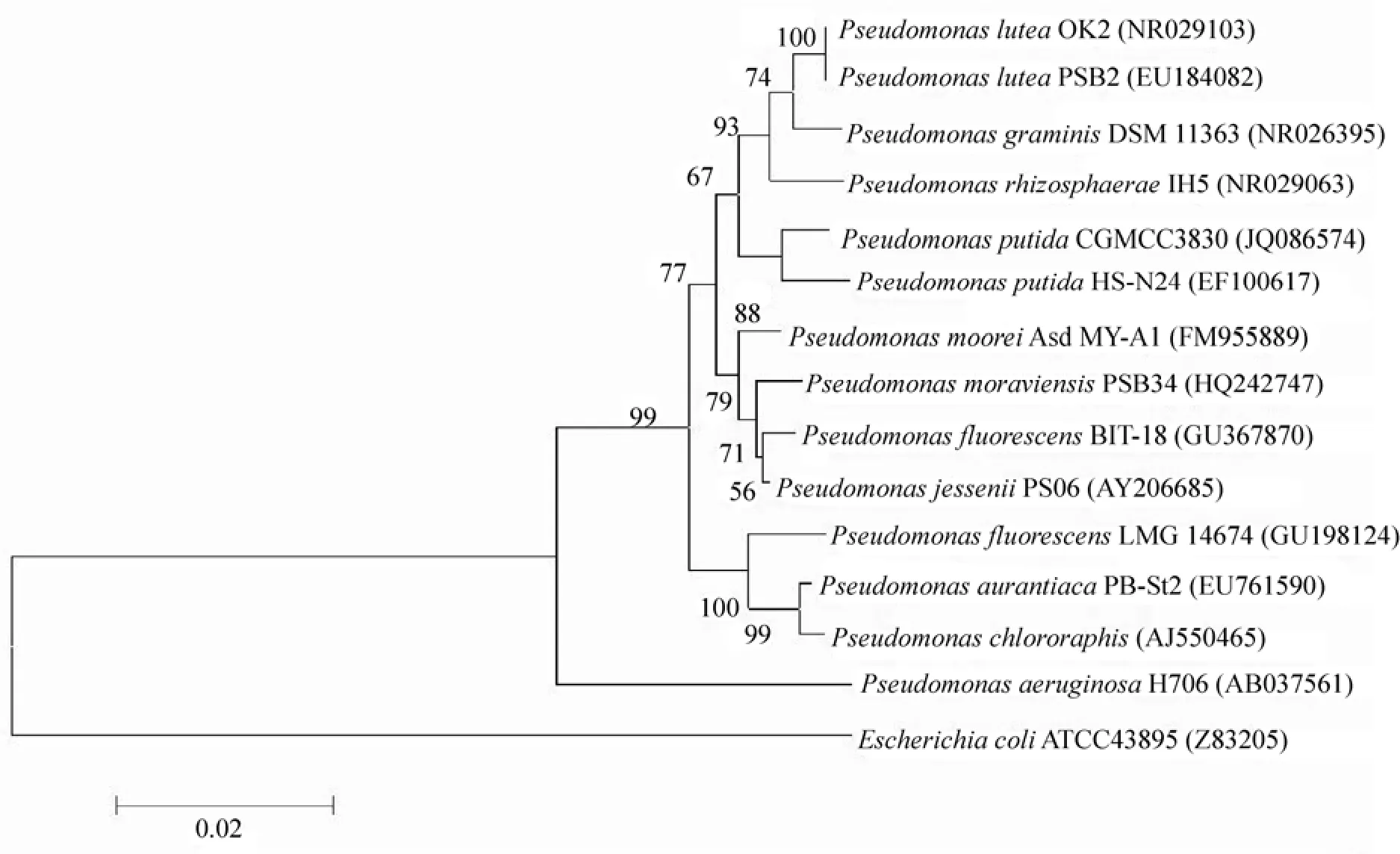
Fig. 1 Phylogenetic tree for P. putida CGMCC3830 and related strains based on the 16S rRNA gene sequence. Numbers after the names of organisms are accession numbers of published sequences. Phylogenetic tree was inferred by using the neighbour-joining methods. The software MEGA 5.02 was used for analysis.
2.3 Effects of nitrogen sources on nitrilase production
Organic and inorganic nitrogen sources (5 g/L) were tested to investigate their effects on the biomass and nitrilase activity of P. putida CGMCC3830 (Fig. 3).
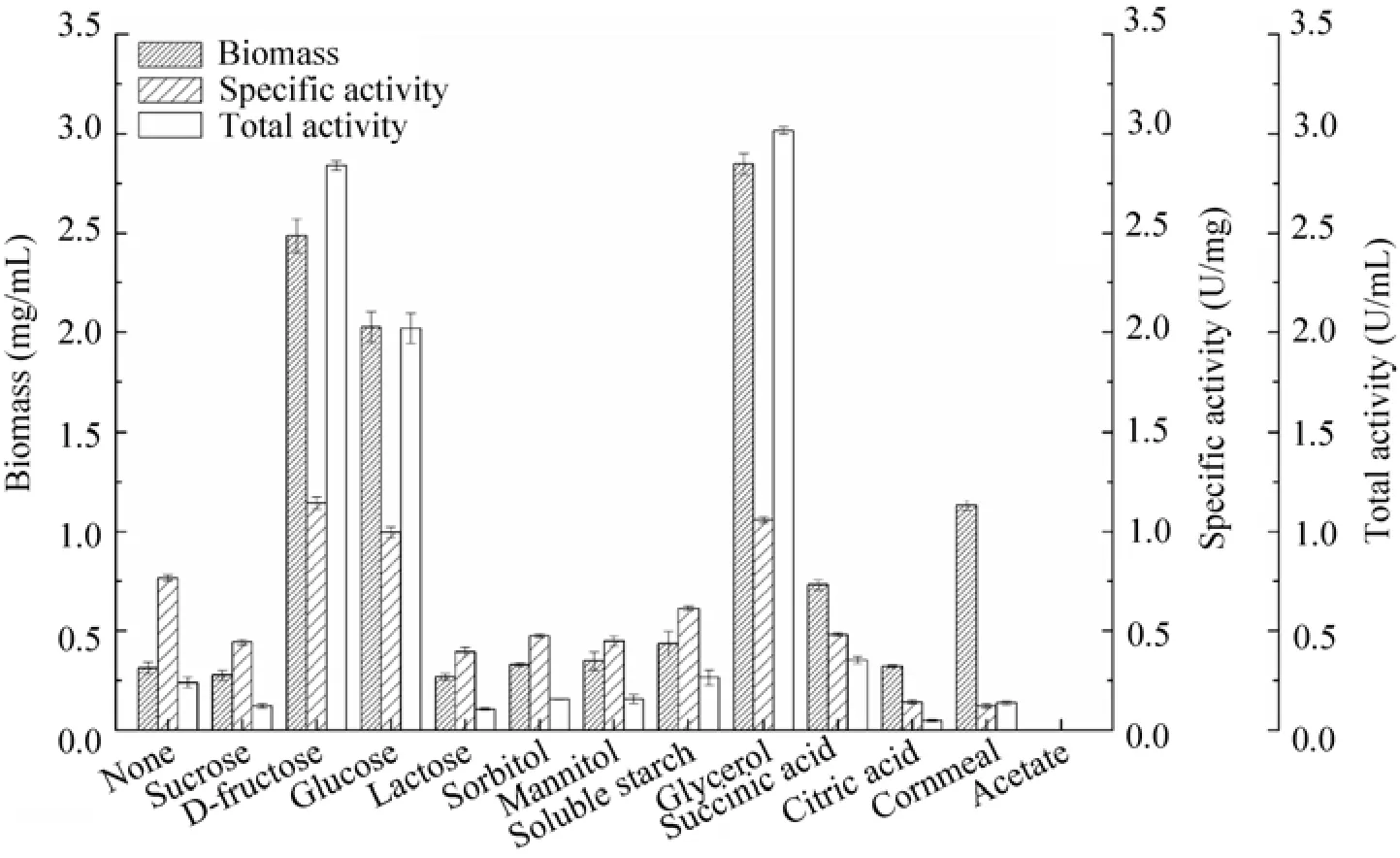
Fig. 2 Effects of carbon sources on biomass and nitrilase activity of P. putida CGMCC3830.
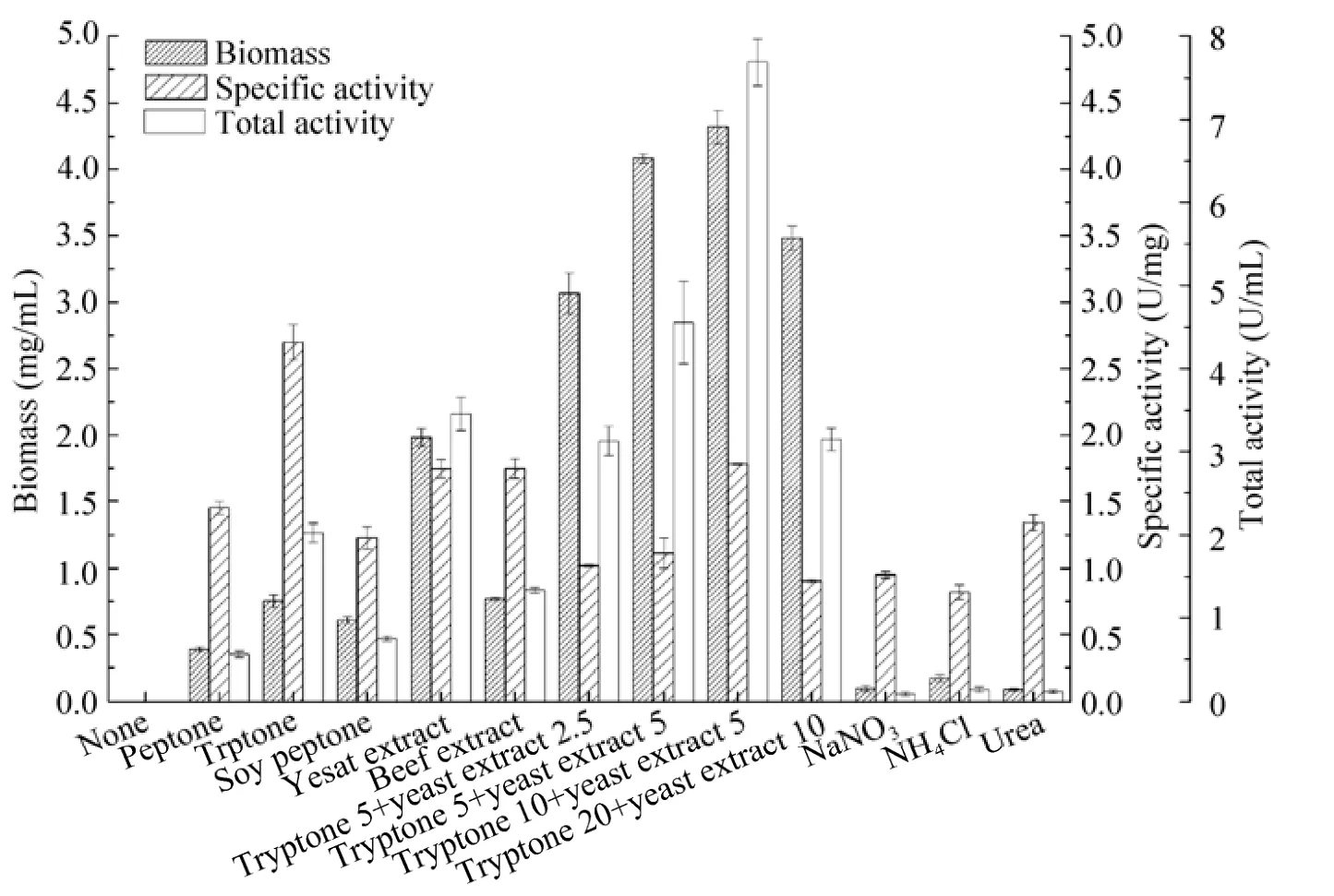
Fig. 3 Effects of nitrogen sources on biomass and nitrilase activity of P. putida CGMCC3830.
The results showed that organic nitrogen sources favored more biomass production compared with inorganic nitrogen sources. Tryptone supported the highest specific activity (2.70 U/mg), but hadrelatively poor biomass (0.75 mg/mL). On the other hand, yeast extract-containing medium supported the most biomass (1.98 mg/mL), but had low specific activity (1.74 U/mg). Therefore, combination of tryptone and yeast extract was further investigated. The combination of 10 g/L tryptone and 5 g/L yeast extract was proven to be the optimum nitrogen source, which enhanced nitrilase production to 7.69 U/mL (biomass 4.32 mg/mL and specific activity 1.78 U/mg). Previous investigations also demonstrated that compound nitrogen sources outperform single nitrogen sources in terms of cell growth and nitrilase formation[28-29].
2.4 Effects of inorganic salts on nitrilase production
Effects of several inorganic salts with final concentration of 0.1 mmol/L on P. putida CGMCC3830 biomass and nitrilase activity were investigated (Fig. 4). The medium with KH2PO4improved the cell growth (biomass 4.21 mg/mL) and nitrilase formation (specific activity 1.89 U/mg) of the species. NaCl did not enhance nitrilase production, but exhibited a beneficial effect on cell growth (biomass 4.32 mg/mL). Nagasawa et al.[30]revealed CuSO4inhibited cell growth of R. rhodochrous J1 and had no effect on nitrilase formation. In this study, CuSO4was proven to strongly inhibit both cell growth (biomass 3.16 mg/mL) and nitrilase formation (specific activity 0.87 U/mg) of P. putida CGMCC3830. Other salts were found to have no obvious effects on biomass and nitrilase activity. Further experiments suggested that the maximal nitrilase production was achieved when KH2PO4and NaCl concentration was 1 g/L.
2.5 Effects of inducers on nitrilase production
Most nitrilases are inducible by certain nitriles, amides, or their analogs[29]. However, appropriate nitrilase activity (specific activity 2.00 U/mg) of P. putida CGMCC3830 was still detected without addition of any inducers. On the other hand, in this study, several nitriles provided negligible benefits on nitrilase expression, except 2-cyanopyridine, which made a slight improvement on specific activity (2.45 U/mg) (Fig. 5). The selected amides and acids were proven to inhibit cell growth, and among them, benzamide also hindered nitrilase formation. The maximal specificactivity (3.38 U/mg) was observed with urea, although urea showed no significant effect on cell growth. Then, a urea concentration of 1 g/L was found to be optimal for nitrilase production.
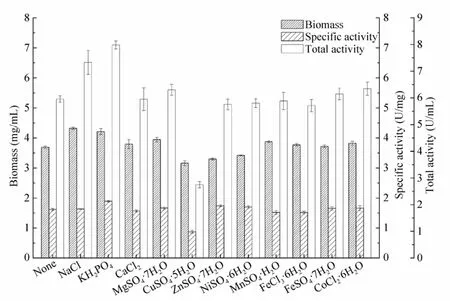
Fig. 4 Effects of inorganic salts on biomass and nitrilase activity of P. putida CGMCC3830.
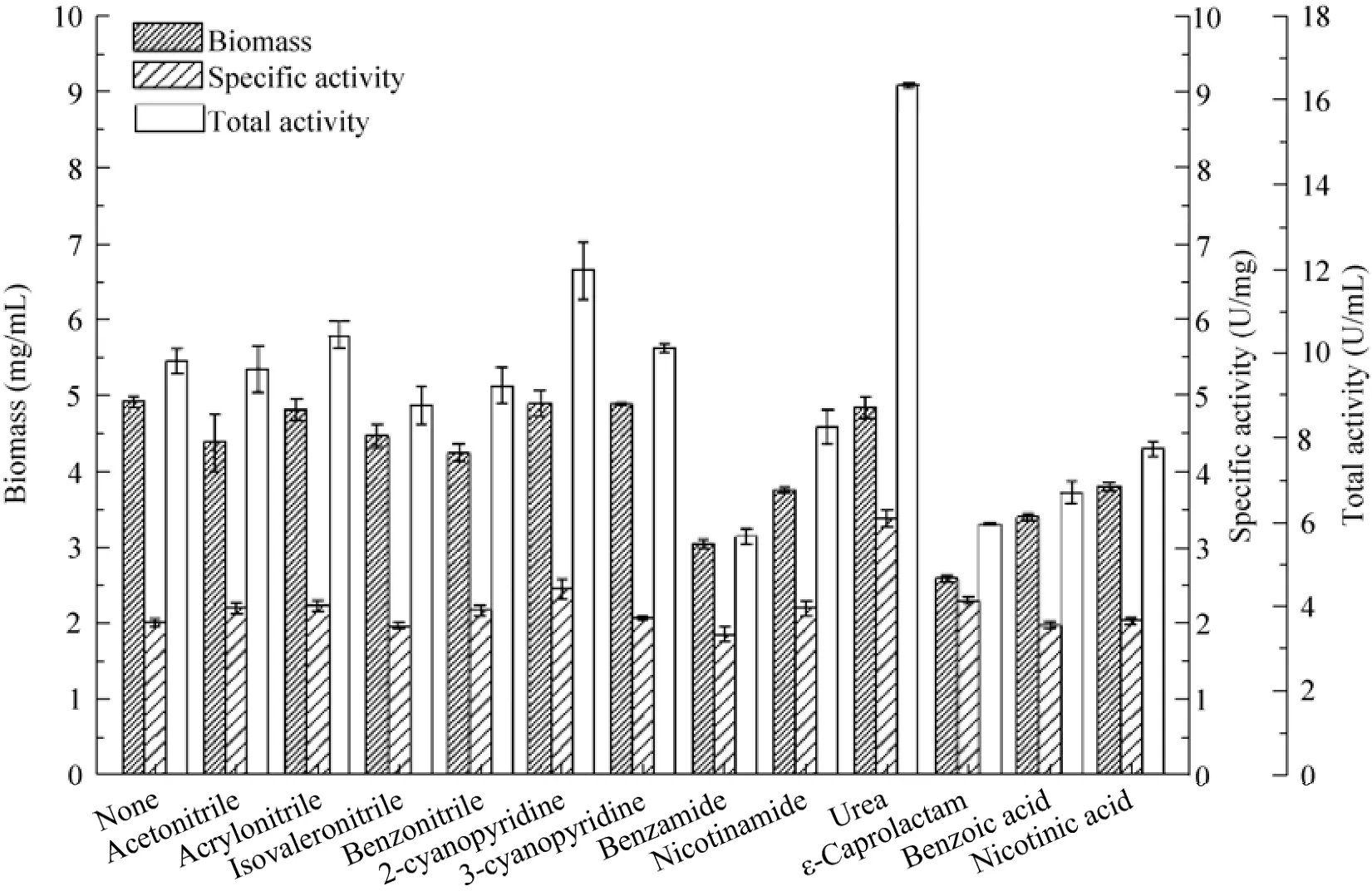
Fig. 5 Effects of inducers on biomass and nitrilase activity of P. putida CGMCC3830.
2.6 Effects of initial pH and culture temperature on nitrilase production
The cultivation conditions including initial pH and culture temperature were also studied for their effects on P. putida CGMCC3830 nitrilase production. The optimal initial pH was 6.0. A sharp drop in total activity was observed when the initial pH was 8.5, which was 31% of the maximum. This strain displayed moderate nitrilase production at temperatures ranging from 25 ℃ to 37 ℃; particularly, the highest nitrilase production was obtained at 30 ℃. Through the cultivation conditions optimization, nitrilase production reached up to 31.34 U/mL.
2.7 Medium composition optimized by RSM
Based on the OFAT experimental results, three variables (glycerol, tryptone, and yeast extract) were chosen for the subsequent RSM experiment. The design and results of the CCD experiments are shown in Table 1. RSM yielded the following regression equation:
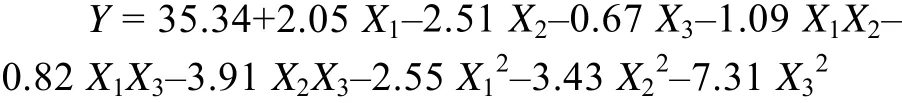
where Y is the total activity (U/mL); and X1, X2, and X3are the coded values of the independent variables glycerol, tryptone, and yeast extract, respectively.
The other culture conditions were fixed based on the results from the OFAT optimization: NaCl 1 g/L, KH2PO41 g/L, urea 1 g/L, initial pH 6.0 and culture temperature 30 ℃.
To validate the regression coefficient, analysis of variance (ANOVA) for nitrilase production was performed (Table 2). The P-value of the model was less than 0.000 1, and the P-value of “lack of fit”was 0.437 0. Thus, the model could adequately fit the experimental data. The model was reliable withan R2value of 0.976 2, thus suggesting that the regression model is suitable for analyzing the response trends. By inspecting the F-values of each factor, tryptone showed the most significant effect on nitrilase production, followed by glycerol and yeast extract. The 3D response surface plots and contour curves show the effects of various culture conditions on total activity (Fig. 6). The contour curves illustrate interactive effect of tryptone and yeast extract on nitrilase production is significant. Substantial interaction was also observed between the other pairs of variables, glycerol and tryptone as well as glycerol and yeast extract.
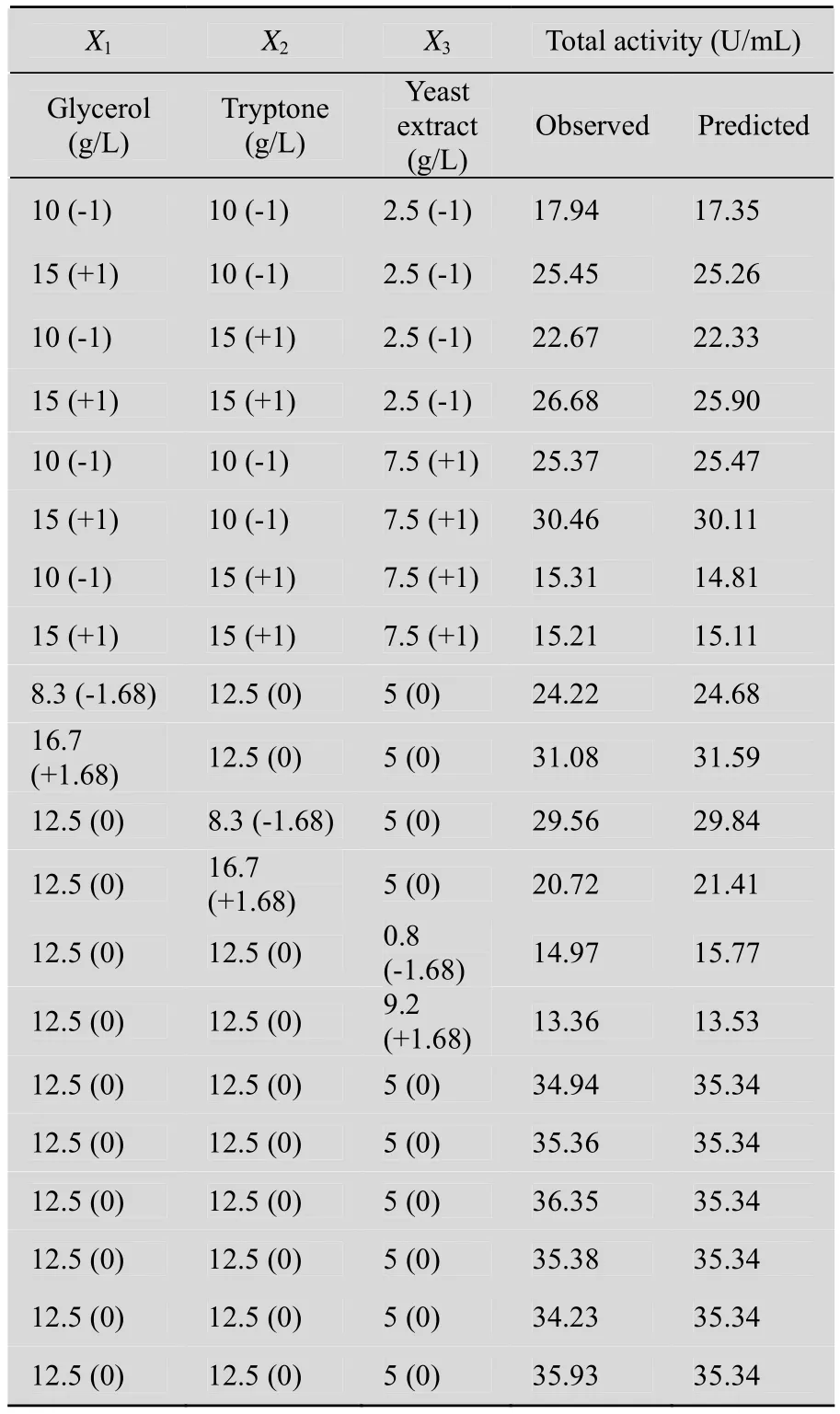
Table 1 Design and results of the central composite design (CCD) experiments
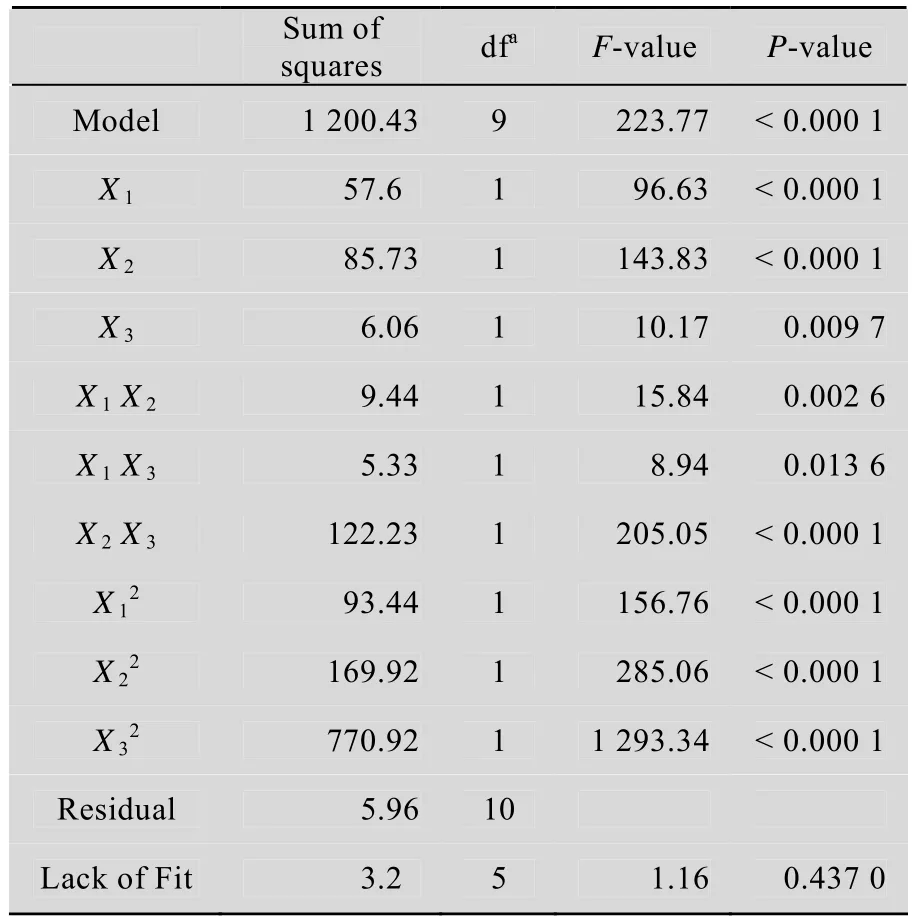
Table 2 Analysis of variance (ANOVA) for the experimental results of CCD
The optimum levels of the investigated factors can be deduced from the 3D response surface plots and the equation obtained from multiple regression analysis. The model predicted that the maximum total activity was located at X1= 13.54 g/L, X2= 11.59 g/L, and X3= 5.21 g/L; the predicted total activity was 36.36 U/mL. The triplicate test shows an actual total activity of 36.12 U/mL, which is close to the predicted response, thus affirming the rationality of the model.
Optimization of culture conditions remains a facile and feasible way to increase enzyme activity and improve the catalytic potential of biocatalyst[11,31]. Through optimization of culture conditions, the total nitrilase activity of P. putida CGMCC3830 was increased from 2.02 U/mL to 36.12 U/mL. Other reported 3-cyanopyridine converting strains, including Rhodobacter sphaeroides LHS-305 and Rhodococcus sp. NDB 1165 showed nitrilase activity of 0.155 and 3.67 U/mL, respectively[32-33].
2.8 Substrate specificity of P. putida CGMCC3830 nitrilase
Nitrilases with different sources are proven tohave diverse substrate specificities for aliphatic nitriles, aromatic nitriles or arylacetonitriles[4,11]. Here, different kinds of nitriles were employed as substrates to investigate the substrate specificity of P. putida CGMCC3830 nitrilase (Table 3). P. putida nitrilase exhibited a broad substrate spectrum and preferentially hydrolyzed aromatic nitriles, particularly 3- and 4-cyanopyridine, followed by benzonitrile, 2-chloro-4-cyanopyridine, and pyrazinecarbonitrile. The aliphatic nitriles such as acrylonitrile, succinonitrile, iminodiacetonitrile and aminoacetonitrile were also somewhat effective substrates, but arylacetonitriles (phenylacetonitrile, mandelonitrile and 4-methoxyphenylacetonitrile) were inert. The genus Pseudomonas generally harbored aliphatic nitrilase or arylacetonitrilase activity[15-19]. In the present study, it was observed that P. putida nitrilase exhibited significant activity towards aromatic nitrile and belonged to aromatic nitrilase.
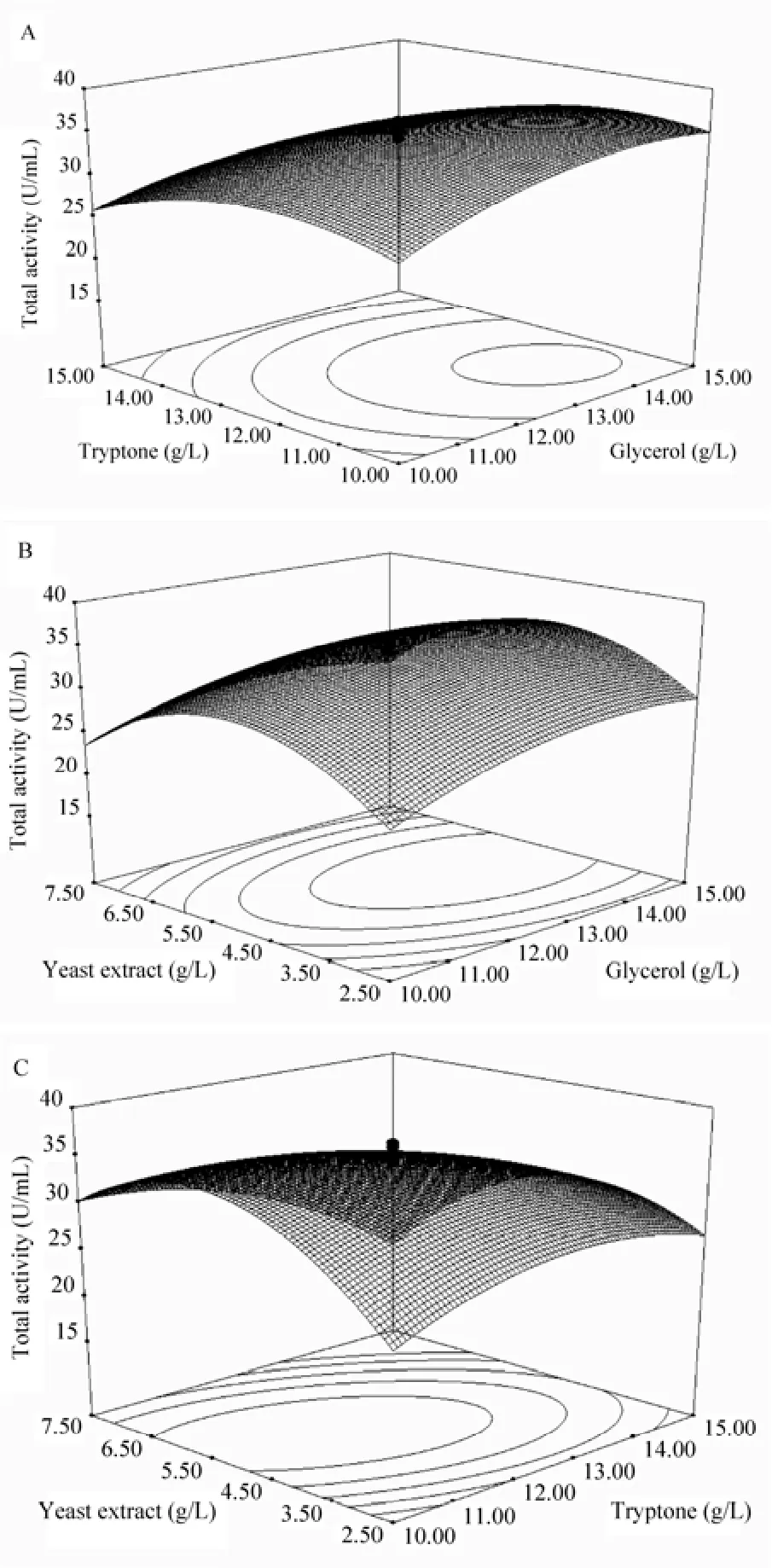
Fig. 6 Response-surface curve of nitrilase production by P. putida CGMCC3830 showing mutual interactions between glycerol and tryptone (A), glycerol and yeast extract (B), tryptone and yeast extract (C). Another variable was fixed at center point level.
2.9 Bioconversion of 3-cyanopyridine into nicotinic acid
P. putida CGMCC3830 resting cells prepared under optimized conditions was applied in nicotinicacid biosynthesis. Bioconversion reaction for 200 mmol/L (20.8 g/L) 3-cyanopyridine was carried out (Fig. 7) in a system of 100 mL potassium phosphate buffer (100 mmol/L, pH 7.2) with
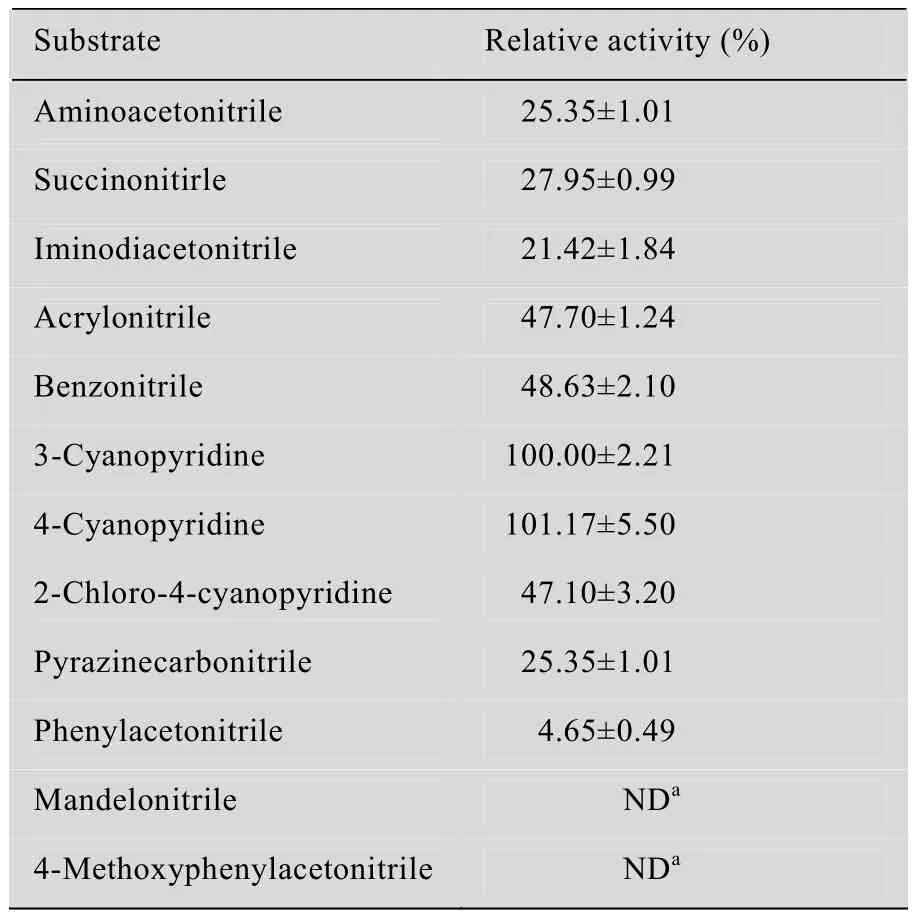
Table 3 Substrate specificity of nitrilase in P. putida CGMCC3830
2 mg/mL of resting cells. The 3-cyanopyridine and nicotinic acid concentrations in the reaction mixture were assayed by HPLC at various intervals. The results showed that 3-cyanopyridine was fully converted within 90 min and corresponding nicotinic acid (about 24.6 g/L) could be obtained, while no amide was detected in the reaction mixture. Prasad and co-workers utilized 2.0 mg/mL Rhodococcus sp. NDB 1165 resting cells for 3-cyanopyridine hydrolysis, and 50 mmol/L (5.2 g/L) 3-cyanopyridine could be completely converted within 20 min at 40 ℃[33]. Recently, Yang et al.[32]reported that R. sphaeroides LHS-305 nitrilase could hydrolyze 200 mmol/L (20.8 g/L) 3-cyanopyridine with 93% conversion rate within 13 h using 6.1 mg/mL of cells at 30 ℃.
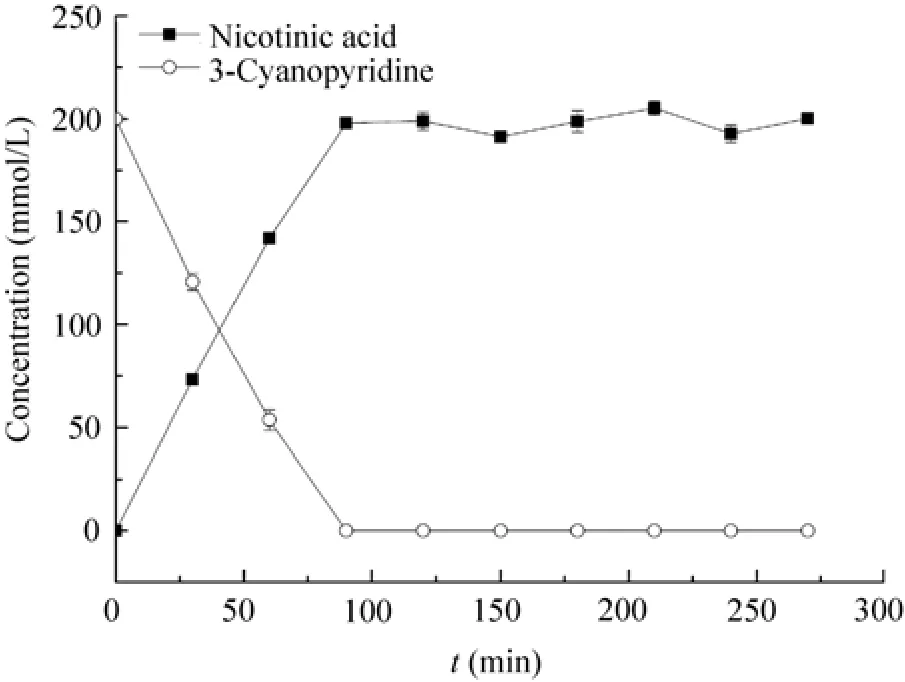
Fig. 7 Time course of nicotinic acid synthesis from 3-cyanopyridine by P. putida CGMCC3830.
3 Conclusions
In this study, a novel bacterium was successfully isolated from soil with 3-cyanopyridine as the sole nitrogen source and proven potential in nitrile hydrolysis. It was identified as P. putida CGMCC3830 according to morphological, physiological, biochemical properties and 16S rRNA gene sequence analysis. The utilization of nitrilase-producing biocatalyst for industrial application required an optimized production strategy. Culture optimization in this study resulted in a significant improvement in total activity from 2.02 U/mL to 36.12 U/mL. The high nitrilase production and the versatile nitrile-converting catalytic property of P. putida CGMCC3830 facilitate its potential application for carboxylic acids synthesis in commercial scale. Further studies including fed-batch reaction and coding gene expression are in progress to fully exploit the biocatalyst for carboxylic acids bioproduction.
REFERENCES
[1] Schmid A, Dordick JS, Hauer B, et al. Industrial biocatalysis today and tomorrow. Nature, 2001, 409: 258–266.
[2] Tao J, Xu JH. Biocatalysis in development of green pharmaceutical processes. Curr Opin Chem Biol, 2009, 13(1): 43–50.
[3] Bornscheuer UT, Huisman GW, Kazlauskas RJ, et al. Engineering the third wave of biocatalysis. Nature, 2012, 485: 185–194.
[4] Thuku RN, Brady D, Benedik MJ, et al. Microbial nitrilases: versatile, spiral forming industrial enzymes. J Appl Microbiol, 2009, 106(3): 703–727.
[5] Sharma NN, Sharma M, Bhalla TC. An improved nitrilase mediated bioprocess for synthesis of nicotinic acid from 3-cyanopyridine with hyperinduced Nocardia globerula NHB-2. J Ind Microbiol Biotechnol, 2011, 38(9): 1235–1243.
[6] Martínková L, Vejvoda V, Kaplan O, et al. Fungal nitrilases as biocatalysts: recent developments. Biotechnol Adv, 2009, 27(6): 661–670.
[7] Velankar H, Clarke KG, Preez R, et al. Developments in nitrile and amide biotransformation processes. Trends Biotechnol, 2010, 28(11): 561–569.
[8] Bhalla TC, Miura A, Wakamoto A, et al. Asymmetric hydrolysis of α-aminonitriles to optically active amino acids by a nitrilase of Rhodococcus rhodochrous PA-34. Appl Microbiol Biotechnol, 1992, 37(2): 184–190.
[9] Brady D, Beeton A, Zeevaart J, et al. Characterization of nitrilase and nitrile hydratase biocatalytic systems. Appl Microbiol Biotechnol, 2004, 64(1): 76–85.
[10] Sharma N, Sharma M, Bhalla T. Nocardia globerula NHB-2 nitrilase catalysed biotransformation of 4-cyanopyridine to isonicotinic acid. AMB Express, 2012, 2: 25.
[11] Gong JS, Lu ZM, Li H, et al. Nitrilases in nitrile biocatalysis: recent progress and forthcoming research. Microb Cell Fact, 2012, 11: 142.
[12] Kumar V, Bhalla TC. Transformation of p-hydroxybenzonitrile to p-hydroxybenzoic acid using nitrilase activity of Gordonia terrae. Biocatal Biotransfor, 2013, 31(1): 42–48.
[13] Naik SC, Kaul P, Barse B, et al. Studies on the production of enantioselective nitrilase in a stirred tank bioreactor by Pseudomonas putida MTCC 5110. Bioresour Technol, 2008, 99(1): 26–31.
[14] Chen J, Zheng RC, Zheng YG, et al. Microbial transformation of nitriles to high-value acids or amides. Adv Biochem Eng Biotechnol, 2009, 113: 33–77.
[15] Robinson WG, Hook RH. Ricinine nitrilase: I. Reaction product and substrate specificity. J Biol Chem, 1964, 239: 4257–4262.
[16] Layh N, Parratt J, Willetts A. Characterization and partial purification of an enantioselective arylacetonitrilase from Pseudomonas fluorescens DSM 7155. J Mol Catal B: Enzym, 1998, 5(5/6): 467–474.
[17] Kiziak C, Conradt D, Stolz A, et al. Nitrilase from Pseudomonas fluorescens EBC191: cloning and heterologous expression of the gene and biochemical characterization of the recombinant enzyme. Microbiology, 2005, 151(11): 3639–3648.
[18] Banerjee A, Kaul P, Banerjee UC. Purification and characterization of an enantioselective arylacetonitrilase from Pseudomonas putida. Arch Microbiol, 2006, 184(6): 407–418.
[19] Kim JS, Tiwari MK, Moon HJ, et al. Identification and characterization of a novel nitrilase from Pseudomonas fluorescens Pf-5. Appl Microbiol Biotechnol, 2009, 83(2): 273–283.
[20] Kennedy M, Krouse D. Strategies for improving fermentation medium performance: a review. J Ind Microbiol Biotechnol, 1999, 23(6): 45–47.
[21] Kammoun R, Naili B, Bejar S. Application of a statistical design to the optimization of parameters and culture medium for α-amylase production by Aspergillus oryzae CBS 819.72 grown on gruel (wheat grinding by-product). Bioresour Technol, 2008, 99(13): 5602–5609.
[22] Ruchi G, Anshu A, Khare SK. Lipase from solvent tolerant Pseudomonas aeruginosa strain: production optimization by response surface methodology and application. Bioresour Technol, 2008, 99(11): 4796–4802.
[23] Holt JG, Kreig NR, Sneath PHA, et al. Bergey’s Manual of Determinative Bacteriology. Williams and Wilkins, Baltimore, MD, 1994.
[24] Thompson JD, Higgins DG, Gibson TJ. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res, 1994, 22(22): 4673–4680.
[25] Fawcett JK, Scott JE. A rapid and precise method for the determination of urea. J Clin Pathol, 1960, 13: 156–159.
[26] Singh R, Sharma R, Tewari N, et al. Nitrilase and its application as a ‘green’ catalyst. Chem Biodivers, 2006, 3(12): 1279–1287.
[27] Banerjee A, Kaul P, Banerjee UC. Enhancing the catalytic potential of nitrilase from Pseudomonas putida for stereoselective nitrile hydrolysis. Appl Microbiol Biotechnol, 2006, 72(1): 77–87.
[28] Hu JG, Wang YJ, Zheng YG, et al. Isolation of glycolonitrile-hydrolyzing microorganism based oncolorimetric reaction. Enzyme Microb Technol, 2007, 41(3): 244–249.
[29] He YC, Xu JH, Su JH, et al. Bioproduction of glycolic acid from glycolonitrile with a new bacterial isolate of Alcaligenes sp. ECU0401. Appl Biochem Biotechnol, 2010, 160(5): 1428–1440.
[30] Nagasawa T, Kobayashi M, Yamada H. Optimum culture conditions for the production of benzonitrilase by Rhodococcus rhodochrous J1. Arch Microbiol, 1988, 150(1): 89–94.
[31] Kim HO, Lim JM, Joo JH, et al. Optimization of submerged culture condition for the production of mycelial biomass and exopolysaccharides by Agrocybe cylindracea. Bioresour Technol, 2005, 96(10):1175–1182.
[32] Yang C, Wang X, Wei D. A new nitrilase-producing strain named Rhodobacter sphaeroides LHS-305: biocatalytic characterization and substrate specificity. Appl Biochem Biotechnol, 2011, 165(7/8): 1556–1567.
[33] Prasad S, Misra A, Jangir V, et al. A propionitrileinduced nitrilase of Rhodococcus sp. NDB 1165 and its application in nicotinic acid synthesis. World J Microbiol Biotechnol, 2007, 23(3): 345–353.
(本文责编 郝丽芳)
《生物工程学报》对摘要的写作要求
1. 研究报告摘要:基本要素包括研究目的、方法、结果和结论 (不用单列标题书写)。目的 (Purpose):主要说明作者写此文章的目的,或说明本文主要要解决的问题;方法 (Methods):重点说明作者的主要工作过程及使用的方法。应用性文章如需要,可注明条件、使用的主要设备和仪器。结果 (Results):本文最后得出的结果 (实验数据部分)。结论 (Conclusions):如系基础研究,应写明本文的创新之处,及文章在讨论部分表述的观点;如系应用性研究,应尽可能提及本文结果和结论的应用范围和应用情况或应用前景。
2. 综述摘要:包括论述内容的发展水平、自己的评论及展望,尤其要注意结合自己的研究工作。
3. 英文摘要的撰写要点:英文摘要的内容应与中文摘要一致,但比中文摘要更详尽。英文摘要完成后,务必请英文较好、且专业知识强的专家审阅定稿后再返回编辑部。凡不符合要求的,即使学术上可以达到刊出的水平,本刊也将推迟发表。
(1) 建议使用第一人称,尽量不使用第三人称和被动语态。
(2) 建议用主动语态,被动语态表达拖拉模糊尽量不用,这样可以免好多长句,以求简单清晰。
(3) 尽量使用清晰简练的短句,避免很长的句子。注意正确使用英文写作习惯和语法。
(4) 摘要应当使用过去时态,语法正确,句子通顺。
(5) 摘要中避免使用缩写语,除非是那些人人皆知的 (如DNA、ATP等),或者确实是非常长,而且出现多次的短语才允许用缩写语,并且在第一次出现时要写出全称。
(6) 在英文摘要中,不要使用任何汉字字符,包括标点、括号、温度、希腊字母等。
(7) 句子的开头处最好不要使用数字。
Screening, identification and culture optimization of a newly isolated aromatic nitrilase-producing bacterium–Pseudomonas putida CGMCC3830
Xiaoyan Zhu1,2, Jinsong Gong1,2, Heng Li1, Zhenming Lu1, Zhemin Zhou2, Jinsong Shi1, and Zhenghong Xu1,2
1 School of Pharmaceutical Science, Jiangnan University, Wuxi 214122, Jiangsu, China
2 The Key Laboratory of Industrial Biotechnology, Ministry of Education, Jiangnan University, Wuxi 214122, Jiangsu, China
Microbial nitrilases have attracted increasing attention in nitrile hydrolysis for carboxylic acid production in recent years. A bacterium with nitrilase activity was isolated and identified as Pseudomonas putida CGMCC3830 based on its morphology, physiological and biochemical characteristics, as well as 16S rRNA gene sequence. The nitrilase production was optimized by varying culture conditions using the one-factor-at-a-time method and response surface methodology. Glycerol 13.54 g/L, tryptone 11.59 g/L, yeast extract 5.21 g/L, KH2PO41 g/L, NaCl 1 g/L, urea 1 g/L, initial pH 6.0 and culture temperature 30 ℃ were proved to be the optimal culture conditions. It resulted in the maximal nitrilase production of 36.12 U/mL from 2.02 U/mL. Investigations on substrate specificity demonstrate P. putida nitrilase preferentially hydrolyze aromatic nitriles. When applied in nicotinic acid synthesis, 2 mg/mL P. putida cells completely hydrolyzed 20.8 g/L 3-cyanopyridine into nicotinic acid in 90 min. The results indicated P. putida CGMCC3830 displayed potential for industrial production of nicotinic acid.
nitrilase, Pseudomonas putida, 3-cyanopyridine, nicotinic acid, optimization
June 4, 2013; Accepted: July 29, 2013
Zhenghong Xu. Tel: +86-510-85918206; E-mail: zhenghxu@jiangnan.edu.cn
朱小燕, 龚劲松, 李恒, 等. 产芳香腈水解酶的恶臭假单胞菌Pseudomonas putida CGMCC3830 的筛选、鉴定及发酵优化. 生物工程学报, 2014, 30(3): 412−424.
Zhu XY, Gong JS, Li H, et al. Screening, identification and culture optimization of a newly isolated aromatic nitrilase-producing bacterium–Pseudomonas putida CGMCC3830. Chin J Biotech, 2014, 30(3): 412−424.
Supported by: National Natural Science Foundation of China (No. 21206055), National High Technology Research and Development Program of China (863 Program) (No. 2011AA02A211).
国家自然科学基金 (No. 21206055),国家高技术研究发展计划 (863计划) (No. 2011AA02A211) 资助。
时间:2013-09-12 网络出版地址:http://www.cnki.net/kcms/detail/11.1998.Q.20130912.0213.002.html

