Differential expression of serum miR-126,miR-141 and miR-21 as novel biomarkers for early detection of liver metastasis in colorectal cancer
Jie Yin,Zhigang Bai,Jianning Song,Yun Yang,Jin Wang,Wei Han,Jun Zhang,Hua Meng,Xuemei Ma,Yao Yang,Tingting Wang,Weirong Li,Zhongtao Zhang
Department of General Surgery,Beijing Friendship Hospital,Capital Medical University,Beijing 100050,China
Introduction
Colorectal cancer (CRC) is one of the most common,lethal diseases.Synchronous liver metastases (SLM) account for approximately 15% of newly diagnosed CRC,which are often resistant to conventional therapies and lead to a poor prognosis (1).Up to 50% of patients with CRC will have colorectal liver metastasis (CLM) at some point (2).Novel biomarkers that are of clinical value are thus in urgent need to improve compliance rates.Recently,a family of small regulatory RNAs,microRNAs (miRNAs),has emerged as possible serum markers for human diseases including cancers,due to their relative stability in the circulation (3).
Evolving evidence suggests that miRNAs play an important role in carcinogenesis and cancer progression through their ability to affect the expression of genes that can regulate the tumorigenic process (4).miRNAs are small non-coding RNAs (18-22 nucleotides in length) that regulate the expression of target genes by interfering with transcription or inhibiting translation (5).Studies have shown that miRNA expression levels differ between normal and tumor tissues and vary among tumor types (6).The implication that they play an important role in oncogenesis has led to the notion of miRNAs as potential biomarkers for the early diagnosis of cancer,predicting prognosis or treatment responses or even as targets in cancer treatment strategies (7).
Studies of miRNAs in CLM have been limited,and most data are from miRNA expression profiles that compared early stage colonic tumors to metastatic CRC(mCRC).Chenget al.(8) demonstrated plasma miR-141 was a biomarker for detecting colon cancer with distant metastasis,and was associated with poor survival.MiRs-21,135a,335,206 and Let-7a could detect the presence of metastases and had a specificity of 87% and a sensitivity of 76% for the presence of metastases (9).However,data for a large cohort of circulating serum sample study of CRC liver metastases have not been reported.The hope is that several liver metastasis-associated miRNAs can be identified by evaluating their expression in well-defined cohorts of early stage CRC.By studying CLM,we expect to identify several miRNAs that allow the earlier detection of CLM.In this study,we evaluated the expression of various metastasis-associated miRNAs in serum samples of localized CRC (L-CRC) and mCRC,including synchronous liver metastasis CRC (SLM-CRC) and other organ-metastasis CRC (OM-CRC).We demonstrated differential expression of 3 of 7 tested miRNAs in CLM.A signature composed of the expression levels of miR-126,miR-141 and miR-21 in the serum significantly correlated with the presence of early stage CRC liver metastases.
Materials and metho ds
Patients and serum samples
Serum samples (N=224) from CRC patients that had been surgically treated were obtained from Beijing Friendship Hospital between 2007 and 2013,including 116 consecutive patients of L-CRC between January 2008 and June 2010,and 108 consecutive patients of mCRC between September 2007 and April 2013.Two cohorts of CRC patients were selected for analysis.The first cohort included 116 serum samples of L-CRC.The second cohort included 108 serum samples of mCRC,including two subgroups,72 with SLM-CRC and 36 with OM-CRC.No patients recruited to this study had received chemotherapy or radiotherapy prior to blood sampling.Venous blood (5 mL) was collected from each patient before surgery and centrifuged at 3,000 r/min for 10 min.Supernatants were recovered and stored at -80 ℃until further analysis.Follow-up data for all recruited patients were acquired and survival time was calculated from the date of surgery to the date of death or last followup on 10 June 2013.The study was approved by the Ethics Review Board of Beijing Friendship Hospital,and written informed consent was obtained from each patient.
RNA extraction and reverse transcription
RNA was isolated using a miRcute miRNA isolation kit(Tiangen,China),following the manufacturer’s protocol for serum/plasma samples with some modification.Briefly,300 μL of human serum was mixed with 300 μL of MZ lysis buffer.After phase separation,the aqueous phase was mixed with ethanol then applied to miRspin and miRelute columns.The miRNAs were eluted with 30 μL of RNasefree water,with 18 μL used for reverse transcription.RNA concentration and purity were assessed using an Eppendorf Biophotometer.RNA concentrations ranged from 26 to 54 ng/μL.Purity of RNA was verified by measuring the absorbances of samples at 260 and 280 nm and determining the 260/280 ratio (acceptable range 1.77-1.92).
Reverse transcription was carried out using an all-inone miRNA fi rst-strand cDNA synthesis kit (Genecopoeia).Final reaction volumes were 25 μL containing 1 μL of 2.5 U/μL poly-A polymerase,1 μL of RTase Mix,5 μL of 5× reaction buffer and 18 μL of purified miRNA.Reverse transcription was performed in a PTC-200 peltier thermal cycle at 37 ℃for 60 min and then 85 ℃ for 5 min.
Detection of serum miRNAs by quantitative PCR (qPCR)
qPCR was conducted on an ABI 7500 instrument in 96-well plates.Each miRNA assay was performed in duplicate with a non-template control contained in each plate.To control for inter-assay variation,samples analyzed on the same plate were for one specific miRNA.We used an allin-one miRNA qPCR kit (Genecopoeia),with 20 μL qPCR mixtures containing 10 μL of 2× all-in-one qPCR Mix,2 μL of all-in-one miRNA qPCR primer,2 μL of universal adaptor primer,0.4 μL of 50× ROX reference dye and 5.6 μL of cDNA.For normalization of sample-to-sample variation during the RNA isolation procedures,syntheticCaenorhabditis elegansmiRNA cel-miR-39 was added to each denatured sample.Amplification was performed on an Applied Biosystems 7500 thermal cycler with a cycling profile of 50 ℃ for 2 min,95 ℃ for 10 min,followed by50 cycles of 95 ℃ for 15 s and 60 ℃ for 1 min.At the end of the 50th cycle,a melt curve analysis was carried out to verify any non-specific amplification.
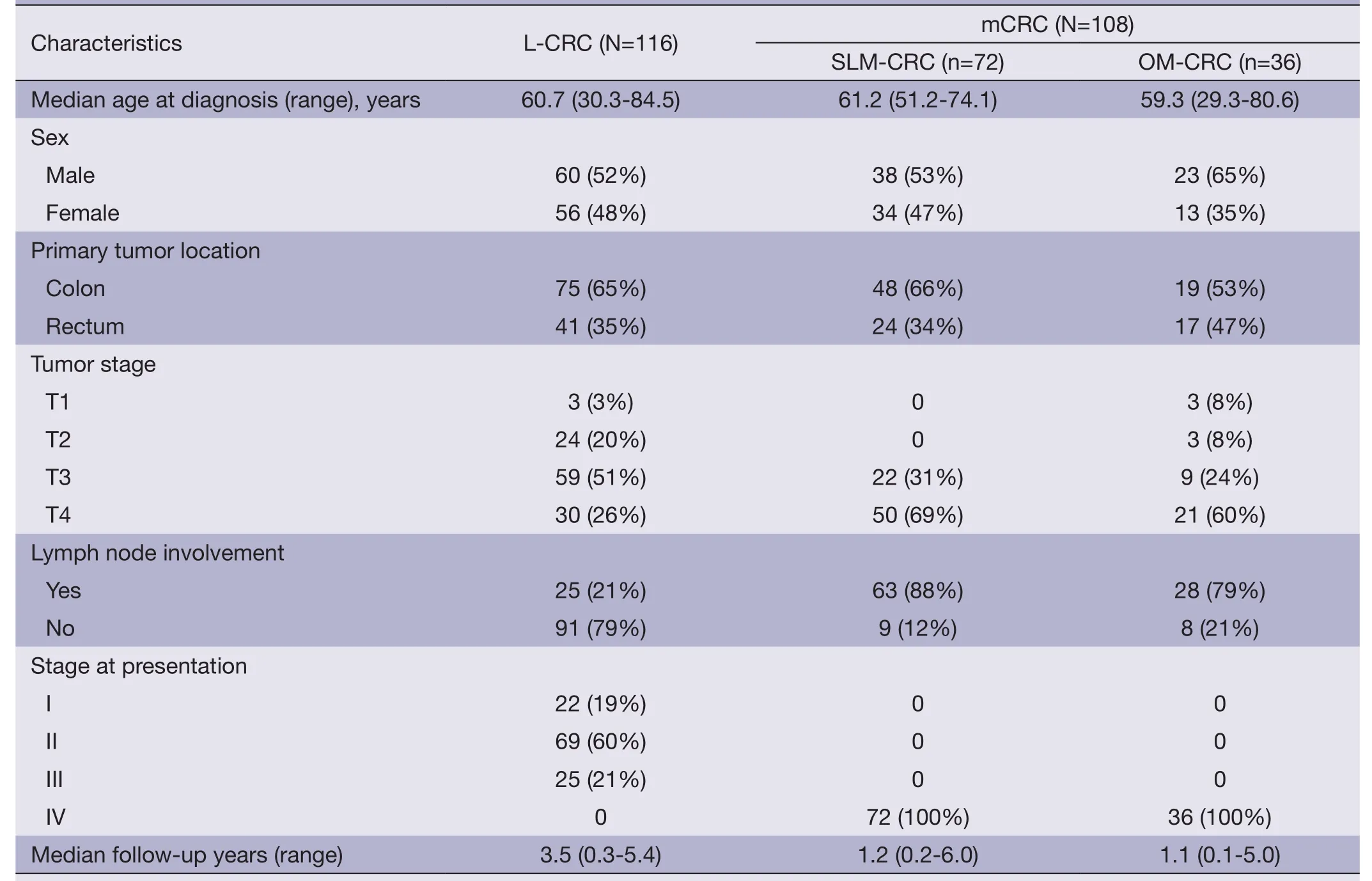
Table 1 Patient characteristics
Statistical analysis
The relative levels of miRNAs were quantified using the 2-ΔΔCtmethod,and the data were analyzed as log10of the relative quantity (log10RQ) of the target miRNAs.Statistical analysis was performed with SPSS version 17.0 (SPSS Inc.,Chicago,IL,USA) and graphs were generated using Graphpad Prism 6.0.Significance of difference between cohorts (L-CRC and mCRC) was determined by two tailed unpairedt-test.A one-way analysis of variance (ANOVA)for variance P value was also performed for the comparison of the L-CRC,SLM-CRC and OM-CRC.Receiver operating characteristic (ROC) curves were generated to assess the diagnostic accuracy of each parameter,and the sensitivity and specificity of the optimum cut-off point were defined as those values that maximized the area under the ROC curve [area under curve (AUC)].All statistical tests were two-sided,and P<0.05 was considered statistically significant.
Results
Patient characteristics
Two cohorts of patient specimens were chosen from the CRC tumor bank at the Beijing Friendship Hospital.Patient characteristics are shown inTable 1.The median age in L-CRC was 60.7 years compared with 61.2/59.3 years in SLM-CRC/OM-CRC,and males were the most common sex in the three subgroups (52%,53% and 65%,respectively).Most of the tumors in the three groups were of colonic origin (65% in L-CRC,66% in SLM-CRC and53% in OM-CRC).L-CRC had a T3 depth of invasion(51%) and the other two subgroups had a T4 depth of invasion (69% in SLM-CRC and 60% in OM-CRC).Of note,a high proportion of patients in SLM-CRC and OM-CRC (88% and 79%) showed mostly lymph node involvement compared with L-CRC (21%).Most patients in L-CRC were at stage II.
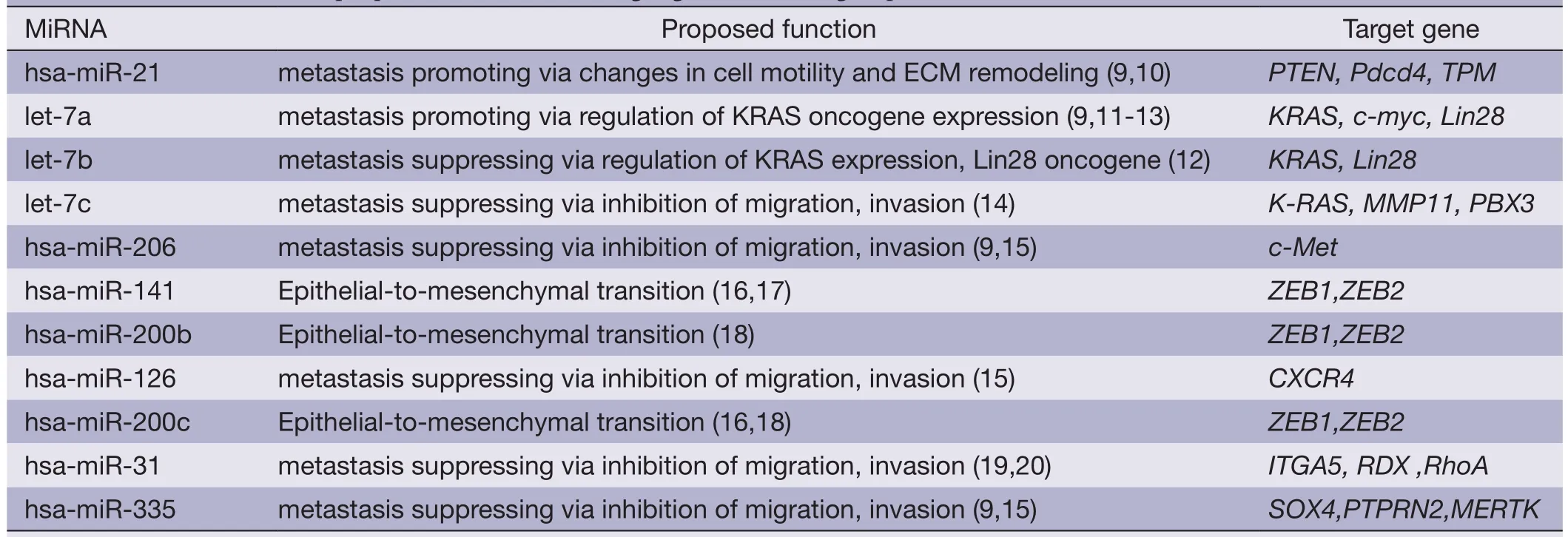
Table 2 MiRNAs and their proposed function with target genes according to published literature
The miRNAs evaluated in this study included miRs-31,335,206,141,126,200b,200c,21,Let7a,Let7b and Let7c that were previously demonstrated to play a role in the metastatic process in a number of tumor models (Table 2).We examined these selected candidate miRNAs by qPCR and only detected the expressions of miRs-31,141,126,21,Let7a,Let7b and Let7c in serum samples,but could not detect miRs-335,206,200b and 200c.
Identification of CRC metastasis-associated miRNAs in serum samples
Statistical analyses to determine significant differences in miRNAs expression between L-CRC and mCRC (SLMCRC and OM-CRC) were performed for all miRNAs evaluated.Among the seven miRNAs that were evaluated,significant changes in expressions were observed for miR-126,Let-7a,miR-141 and miR-21 in mCRC compared with L-CRC (Figure 1).
MiR-126 levels were significantly down-regulated in mCRC when compared with L-CRC (mean log10RQ: -0.3980vs.-0.0204,P<0.0001).But,Let-7a expression levels were significantly up-regulated in mCRC compared with L-CRC (mean log10RQ: -0.1076vs.-0.2747,P=0.0120).Similarly,both serum levels of miR-21 and miR-141 could significantly up-regulated in mCRC from L-CRC by statistical analysis (mean log10RQ: 0.1026vs.-0.3655,P<0.0001; 0.1625vs.-0.4976,P<0.0001).In contrast,serum levels of Let-7b,miR-31and Let-7c did not show significant differences between mCRC and L-CRC (mean log10RQ: -0.2321vs.-0.2617,P=0.7133; -0.3779vs.-0.2925,P=0.2517; -0.3289vs.-0.2944,P=0.6830).Therefore,based on these observations,miR-126,Let-7a,miR-21 and miR-141 were identified as metastasis-associated miRNAs.
Differential expression of miR-126,miR-141 and miR-21 in CRC with liver metastasis
To identify CRC liver metastatic-associated miRNAs in serum samples,we divided the 108 serum samples from mCRC into 72 SLM-CRC and 36 OM-CRC.Among the seven miRNAs that were evaluated,significant changes in expression were observed for miR-126,miR-21 and miR-141 in SLM-CRC compared with L-CRC (Figure 2).
Similar to metastasis-associated miRNAs,miR-126 levels were significantly down-regulated in SLM-CRC and OM-CRC compared with L-CRC (mean log10RQ: -0.4610vs.-0.0204,P<0.0001; -0.2720vs.-0.0204,P=0.0165),but miR-126 expression levels were not significantly different when SLMCRC was compared with OM-CRC (Figure 2A).MiR-141 levels were significantly up-regulated in SLM-CRC and OM-CRC compared with L-CRC (mean log10RQ: 0.0726vs.-0.4976,P<0.0001; 0.3422vs.-0.4976,P<0.0001),and miR-141 expression levels were also significantly different when SLM-CRC was compared with OM-CRC (mean log10RQ: 0.0726vs.0.3422,P=0.0100) (Figure 2B).Similarly,miR-21 levels were significantly up-regulated in SLM-CRC and OM-CRC compared with L-CRC (mean log10RQ: 0.1005vs.-0.3655,P<0.0001; 0.1068vs.-0.3655,P<0.0001),but miR-21 expression levels were not significantly different when SLM-CRC was compared with OM-CRC (Figure 2C).Of interest,Let-7a levels were not significantly different in SLM-CRC compared with L-CRC (mean log10RQ: -0.2036vs.-0.2747,P=0.3203),but were significantly up-regulated in OM-CRC when compared with L-CRC and SLM-CRC(mean log10RQ: 0.0843vs.-0.2747,P<0.0001; 0.0843vs.-0.2036,P=0.0083) (Figure 2D).The other three miRNAs miR-31,Let-7b and Let-7c were not significantly different when SLM-CRC was compared with L-CRC.Therefore,miR-126,miR-21 and miR-141 were proposed as CRC liver metastasis-associated miRNAs.
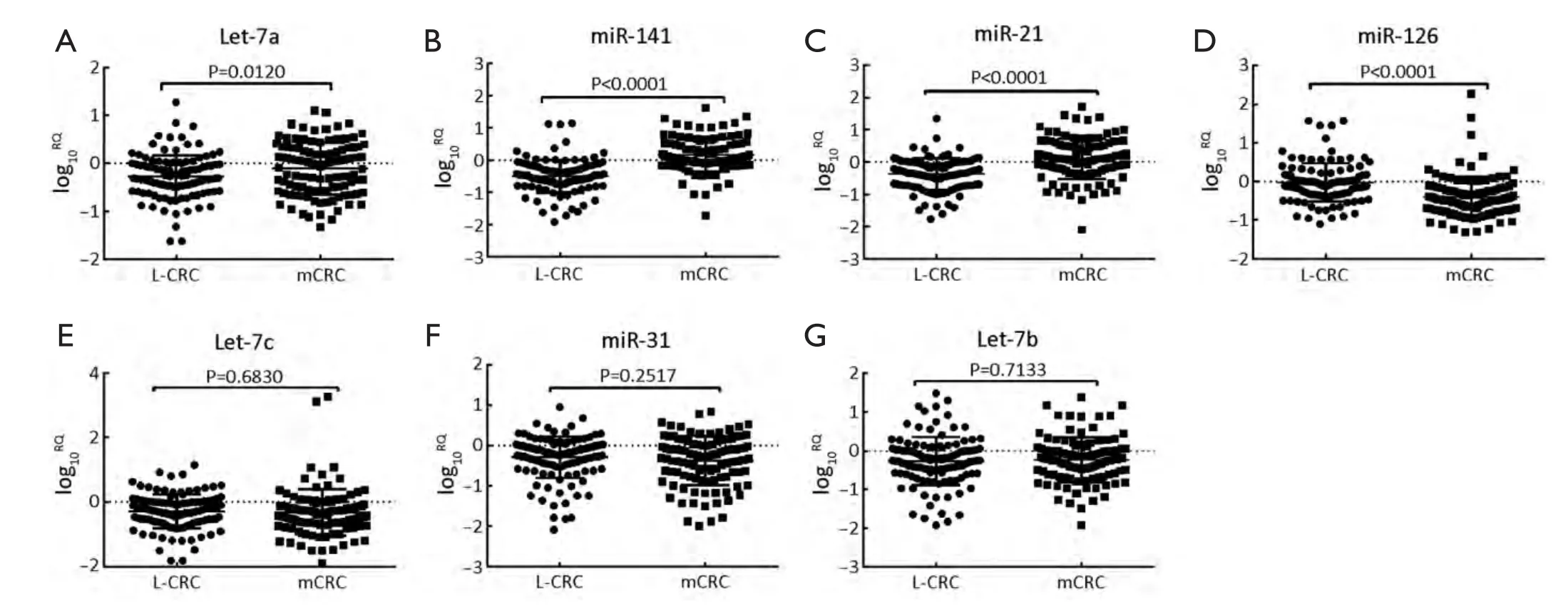
Figure 1 Serum microRNA (miRNA) levels in localized colorectal cancer (L-CRC) and metastatic CRC (mCRC).(A-D) Compared with L-CRC,serum Let-7a,miR-141 and miR-21 were significantly up-regulated in mCRC,and miR-126 levels were significantly downregulated in mCRC; (E-G) Serum levels of Let-7c,miR-31 and Let-7b did not show significant differences between mCRC and L-CRC.mCRC includes synchronous liver-metastatic colorectal cancer (SLM-CRC) and organ-metastatic colorectal cancer (OM-CRC).
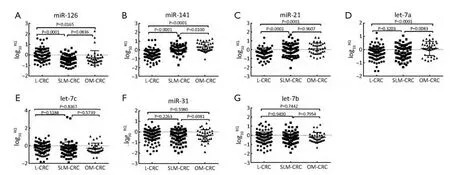
Figure 2 Serum microRNA (miRNA) levels in CRC liver metastases.(A-C) Serum miR-126 levels were significantly down-regulated in synchronous liver-metastatic colorectal cancer (SLM-CRC) and organ-metastatic colorectal cancer (OM-CRC) when compared with localized colorectal cancer (L-CRC).MiR-141 and miR-21 levels were significantly up-regulated in SLM-CRC and OM-CRC when compared with L-CRC; (D) Let-7a levels were up-regulated in OM-CRC in comparison with L-CRC; (E-G) Serum levels of Let-7c,miR-31 and Let-7b did not show significant differences in these three subgroups.
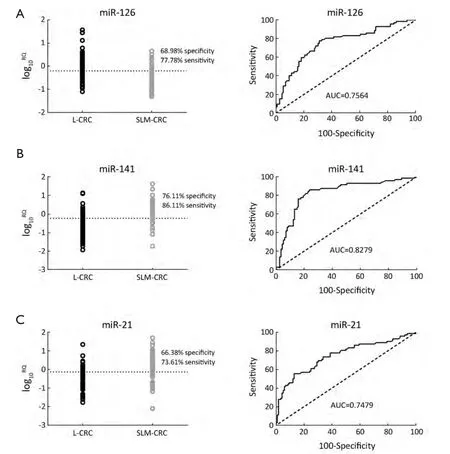
Figure 3 Serum microRNAs (miRNAs) were potential early diagnosis signature for colorectal cancer (CRC) liver metastates.(A) Serum miR-126 had a cut-off with 77.78% sensitivity and 68.97% specificity with an area under curve (AUC) of 0.7564; (B) Serum miR-141 had a cut-off with 86.11% sensitivity and 76.11% specificity with an AUC of 0.8279; (C) Serum miR-21 had a cut-off with 73.61% sensitivity and 66.38% specificity with an AUC of 0.7479.
Potential miRNAs as an early diagnosis signature for CRC liver metastatic disease
These results demonstrated that among the seven selected miRNAs,miR-126,miR-141 and miR-21 were significantly correlated with CRC liver metastasis.The ROC curve was plotted to identify a cut-off value that could distinguish SLM-CRC from L-CRC.The scatter plot showed that at the optimal cut-off,serum miR-126 had a cut-off(log10RQ=-0.2005) with 77.78% sensitivity and 68.97%specificity with an AUC of 0.7564 (Figure 3A),serum miR-141 had a cut-off (log10RQ=-0.2285) with 86.11% sensitivity and 76.11% specificity with an AUC of 0.8279 (Figure 3B),and serum miR-21 had a cut-off (log10RQ=-0.1310) with 73.61% sensitivity and 66.38% specificity with an AUC of 0.7479 (Figure 3C).These results suggest that serum miR-126,miR-141 and miR-21 might be a novel early diagnosis signature for CRC liver metastatic disease.
Discussion
In the present study,using patient serum,we showed that previously suggested metastasis-associated miRNAs (21)are also important in the progression and metastatic potential of CRCs.We showed the differential expression of miR-126,miR-141 and miR-21 during the progression of liver mCRC.Identification of miRNA prognostic signature suggests that miRNA profiles may compliment clinicopathologic variables in predicting the risk of liver metastasis in resected colorectal tumors.
We examined selected candidate miRNAs by qPCR and only detected the expression of 7 of 11 tested miRNAs in serum samples.miRs-335,206,200b and 200c were not detected.The most robust data regarding these 4 miRNA expression profiles were reported by researches using tumor tissues or cells lines,but few have used serum samples(9,15,16,18).Therefore,miR-335,206,200b and 200c may not be secretory miRNAs,which may explain why they were not observed in serum of CRC patients.
Our findings fail to confirm that miR-31,Let-7b and Let-7c are CRC metastasis-associated miRNAs.Previous studies showed that expression of miR-31 inhibited breast cancer metastasis in tumor cells via regulation of a cohort of metastasis-associated genes,includingITGA5,RDX,andRhoA(19,20).Let-7b can suppress the expression of tumor promoter geneLIN28B,which promotes cell migration and invasion,and transforms immortalized colonic epithelial cells (12).Hanet al.(14) demonstrated that Let-7c,in addition to its role in tumor growth suppression,could also function as a tumor metastasis suppressor in CRC by directly destabilizing the mRNAs ofMMP11andPBX3.Our data were inconsistent with these reports,possibly because our research samples were serum from CRC patients,adding further evidence that there are significant differences betweenin vitroandin vivostudies.Further validation in a larger cohort of samples is needed to determine conclusively whether miR-31,Let-7b and Let-7c serve as circulating markers for CRC metastasis-associated miRNAs.
Let-7a is expressed at higher than normal levels in many cancers including CRC,and plays a role in the inhibition of tumor growth by targeting oncogenes such asKRAS(22) andNIRF,and several cell cycle-related genes (23).Experimental and early clinical data revealed Let-7 had these effects and had up-regulating expression in the metastatic disease,especially in the presence ofKRASmutations.Vickerset al.showed elevated expression in metastatic disease compared to normal mucosa or non-metastatic disease,and only inKRASmutation positive tumors (9).In the present study,we found that serum levels of Let-7a were significantly up-regulated in mCRC and OM-CRC when compared with L-CRC (P=0.0120 and P<0.0001,respectively).One limitation of our study was not evaluating theKRASmutation status in tumors,and the failure to gain further insight into the molecular mechanisms and the correlation between Let-7a andKRAS.
The tumor suppression genesPTEN,Pdcd4and tropomyosin 1 (TPM1) were suggested as targets of miR-21 (9,24,25).Clinically,higher miR-21 expression increased with tumor metastatic potential by evaluating the expression of target miRNAs in micro-dissected paraffin embedded CRC tumors,adjacent normal tissue and corresponding liver metastatic tissues (9).Toiyamaet al.found that serum miR-21 was a promising biomarker for the early detection and prognosis of CRC when compared with CRC patients,advanced adenoma and control subjects (26).However in contrast to this previous study that analyzed miRNAs isolated from stage IV,we evaluated miRNA expression specifically in SLM-CRC and OM-CRC.Our findings validate that miR-21 levels were significantly up-regulated in mCRC,SLM-CRC and OM-CRC compared with L-CRC.So serum miR-21 was significantly correlated with CRC liver metastasis.
MiR-126 is essential in angiogenesis by modulating vascular endothelial growth factor receptor (VEGFR 2)related signal transduction through the RAS/ERK and PI3K/AKT pathways by inhibiting regulatory units.High levels of miR-126 in CRC are associated with high VEGFR-2 mRNA and protein levels and a higher density of newly formed microvessels (27).As a metastasis suppressor miRNA,miR-126 was originally defined in breast cancer (15).Zhanget al.described a previously unrecognized role of miR-126 and miR-126*in inhibiting breast cancer metastasis by impeding cytokine-dependent infiltration of two important metastasis-promoting stromal cell types into the primary tumor (28).Recently,miR-126 was shown to suppress metastatic endothelial recruitment and angiogenesis at the site of metastatic colonization by coordinating the targeting of pro-angiogenicIGFBP2,PITPNC1andMERTKgenes (29).Kanget al.demonstrated that both strands of the miR-126 duplex could inhibit lung metastasis to reduce the recruitment of mesenchymal stem cells and inflammatory monocytes to primary tumours (30).Consistent with these reports,we present further evidence that miR-126 levels were significantly down-regulated in mCRC,SLM-CRC and OM-CRC when compared with L-CRC (P<0.0001,P<0.0001 and P=0.0165,respectively).Schepeleret al.(31) demonstrated that miRNA-126 was up-regulated in the primary tumor tissue from patients with stage II microsatellite stable (MSS) colon cancers who experienced relapse significantly later compared with those who did not.That study used tumor tissue samples and only included MSS.The results of miR-126 were obtained from miRNA microarray expression profiles,which failed to amplify miRNA expression by real-time reverse transcription-PCR.However,the underlying mechanisms of its decrease in mCRC require further investigation.
MiR-141 belongs to the miR-200 family,and functions as a switch that can regulate epithelial-to-mesenchymal transition (EMT) and mesenchymal-to-epithelial transition (MET) in human CRC metastasis (16),which promotes MET by inhibiting the expression of E-cadherin transcriptional suppressorsZEB1andZEB2(17).In this study,we found miR-141 was up-regulated in mCRC,SLM-CRC and OM-CRC (P<0.0001,P<0.0001 and P<0.0001,respectively) by quantitative real-time PCR(qRT-PCR) when compared with L-CRC.Chenget al.found that plasma miR-141 was significantly elevated in stage IV cases when compared with stage I-II,stage III and stage I-III combined (8),which was consistent with the results of our study.Huret al.demonstrated that liver metastasis tissues showed higher expression of miR-141 and miR-200c compared with primary CRC (16).However,the liver metastases tissues were not matched to primary tumors and the miR-141 data from circulating serum were not reported.The concentration of miRNAs is not equal in different sources of samples,such as serum and tissue.In our study,miR-141 expression levels were also significantly different when SLM-CRC was compared with OMCRC (P=0.0100),increased serum miR-141 might reflect increased miR-141 levels in the primary tissue and newly formed metastatic foci,and different organs’ metastatic foci maybe induce different miRNAs levels.This hypothesis will need to be tested with circulating serum and matching primary tumors and metastases in the future.
In conclusion,serum miR-126,miR-141 and miR-21 levels in liver mCRC were significantly differential expression compared with L-CRC,suggesting serum miR-126,miR-141 and miR-21 may be novel biomarkers for the clinical diagnosis of early stage liver mCRC,but whether they are biomarkers of liver-specific expression,the expressions of these miRNAs and their downstream target genes in pairs of primary CRC and corresponding liver metastasis tissue specimens need to be analyzed.
Acknowledgements
Disclosure:The authors declare no conflict of interest.
1.Manfredi S,Lepage C,Hatem C,et al.Epidemiology and management of liver metastases from colorectal cancer.Ann Surg 2006;244:254-9.
2.Aldrighetti L,Castoldi R,Di Palo S,et al.Prognostic factors for long-term outcome of hepatic resection for colorectal liver metastases.Chir Ital (In Italian)2005;57:555-70.
3.Cortez MA,Calin GA.MicroRNA identification in plasma and serum: a new tool to diagnose and monitor diseases.Expert Opin Biol Ther 2009;9:703-11.
4.Iorio MV,Croce CM.MicroRNAs in cancer:small molecules with a huge impact.J Clin Oncol 2009;27:5848-56.
5.Fabbri M,Croce CM,Calin GA.MicroRNAs.Cancer J 2008;14:1-6.
6.Lu J,Getz G,Miska EA,et al.MicroRNA expression profiles classify human cancers.Nature 2005;435:834-8.
7.Esquela-Kerscher A,Slack FJ.Oncomirs-microRNAs with a role in cancer.Nat Rev Cancer 2006;6:259-69.
8.Cheng H,Zhang L,Cogdell DE,et al.Circulating plasma miR-141 is a novel biomarker for metastatic colon cancer and predicts poor prognosis.PLoS One 2011;6:e17745.
9.Vickers MM,Bar J,Gorn-Hondermann I,et al.Stagedependent differential expression of microRNAs in colorectal cancer: potential role as markers of metastatic disease.Clin Exp Metastasis 2012;29:123-32.
10.Schetter AJ,Leung SY,Sohn JJ,et al.MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma.JAMA 2008;299:425-36.
11.Zhang W,Winder T,Ning Y,et al.A let-7 microRNA-binding site polymorphism in 3'-untranslated region of KRAS gene predicts response in wild-type KRAS patients with metastatic colorectal cancer treated with cetuximab monotherapy.Ann Oncol 2011;22:104-9.
12.King CE,Wang L,Winograd R,et al.LIN28B fosters colon cancer migration,invasion,and transformation through let-7 dependent and independent mechanisms.Oncogene 2011;30:4185-93.
13.Akao Y,Nakagawa Y.Let-7 microRNA functions as a potential growth suppressor in human colon cancer cells.Biol Pharm Bull 2006;29:903-6.
14.Han HB,Gu J,Zuo HJ,et al.Let-7c functions as a metastasis suppressor by targeting MMP11 and PBX3 in colorectal cancer.J Pathol 2012;226:544-55.
15.Tavazoie SF,Alarcón C,Oskarsson T,et al.Endogenous human microRNAs that suppress breast cancer metastasis.Nature 2008;451:147-52.
16.Hur K,Toiyama Y,Takahashi M,et al.MicroRNA-200c modulates epithelial-tomesenchymal transition (EMT) in human colorectal cancer metastasis.Gut 2013;62:1315-26.
17.Korpal M,Lee ES,Hu G,et al.The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2.J Biol Chem 2008;283:14910-4.
18.Gibbons DL,Lin W,Creighton CJ,et al.Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression.Genes Dev 2009;23:2140-51.
19.Valastyan S,Reinhardt F,Benaich N,et al.A Pleiotropically Acting MicroRNA,miR-31,Inhibits Breast Cancer Metastasis.Cell 2009;137:1032-46.
20.Valastyan S,Benaich N,Chang A,et al.Concomitant suppression of three target genes can explain the impact of a microRNA on metastasis.Genes Dev 2009;23:2592-7.
21.Hurst DR,Edmonds MD,Welch DR.Metastamir: the fi eld of metastasis-regulatory microRNA is spreading.Cancer Res 2009;69:7495-8.
22.Ruzzo A,Graziano F,Vincenzi B,et al.High let-7a microRNA levels in KRAS-mutated colorectal carcinomas may rescue anti-EGFR therapy effects in patients with chemotherapy-refractory metastatic disease.Oncologist 2012;17:823-9.
23.Wang F,Zhang P,Ma Y,et al.NIRF is frequently upregulated in colorectal cancer and its oncogenicity can be suppressed by let-7a microRNA.Cancer Lett 2012;314:223-31.
24.Asangani IA,Rasheed SA,Nikolova DA,et al.MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion,intravasation and metastasis in colorectal cancer.Oncogene 2008;27:2128-36.
25.Zhu S,Si ML,Wu H,et al.MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1).J Biol Chem 2007;282:14328-36..
26.Toiyama Y,Takahashi M,Hur K,et al.Serum miR-21 as a Diagnostic and Prognostic Biomarker in Colorectal Cancer.J Natl Cancer Inst 2013;105:849-59.
27.Hansen TF,Andersen CL,Nielsen BS,et al.Elevated microRNA-126 is associated with high vascular endothelial growth factor receptor 2 expression levels and high microvessel density in colorectal cancer.Oncol Lett 2011;2:1101-6.
28.Zhang Y,Yang P,Sun T,et al.miR-126 and miR-126*repress recruitment of mesenchymal stem cells and inflammatory monocytes to inhibit breast cancer metastasis.Nat Cell Biol 2013;15:284-94.
29.Png KJ,Halberg N,Yoshida M,et al.A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells.Nature 2011;481:190-4.
30.Ren G,Kang Y.A one-two punch of miR-126/126*against metastasis.Nat Cell Biol 2013;15:231-3.
31.Schepeler T,Reinert JT,Ostenfeld MS,et al.Diagnostic and prognostic microRNAs in stage II colon cancer.Cancer Res 2008;68:6416-24.
 Chinese Journal of Cancer Research2014年1期
Chinese Journal of Cancer Research2014年1期
- Chinese Journal of Cancer Research的其它文章
- Prediction rule for estimating advanced colorectal neoplasm risk in average-risk populations in southern Jiangsu Province
- Long-term fatigue state in postoperative patients with breast cancer
- Diffusion-weighted images (DWI) without ADC values in assessment of small focal nodules in cirrhotic liver
- Annual report on status of cancer in China,2010
- EGFR gene copy number as a predictive biomarker for resistance to anti-EGFR monoclonal antibodies in metastatic colorectal cancer treatment: a meta-analysis
- Overexpression of HOXB9 promotes metastasis and indicates poor prognosis in colon cancer
