Characteristics of the gastrointestinal microbiome in children with autism spectrum disorder: a systematic review
Xinyi CAO, Ping LIN, Ping JIANG, Chunbo LI*
•Systematic review•
Characteristics of the gastrointestinal microbiome in children with autism spectrum disorder: a systematic review
Xinyi CAO, Ping LIN, Ping JIANG, Chunbo LI*
1. Introduction
Autism spectrum disorders (ASD) refers to a range of neurodevelopmental disorders, primarily characterized by difficulties in social interactions, verbal and nonverbal communication, and stereotypic or repetitive behaviors.[1]Although the classification of ASD varies in different diagnostic systems, it essentially encompasses autism, childhood disintegrative disorder, Asperger’s syndrome and pervasive developmental disorder not otherwise specified (PDD-NOS).[2,3,4]The etiological pathways and pathogenesis of ASD remain unknown.
A high prevalence (23-70%) of Gastrointestinal(GI) distress, including abdominal pain, bloating,diarrhea and constipation, has been reported among children with ASD.[5]The high frequencies of these GI symptoms could reflect abnormal GI microflora in ASD children. There are more than 100 trillion symbiotic microorganisms[6]in the human GI tract that are important for nutrition and metabolic processes. Some evidence suggests that these same microorganisms also play a role in brain development, behavior, and gene expression via neural, endocrine, and immune pathways.[7]Emerging research on the gut-microbiomebrain connection in both mice and humans has shed new light on the pathogenesis of various mental diseases including ASD.[8,9]Nonetheless, whether GI microbiota contribute to the pathogenesis or the developmental course of ASD is still unclear. Previous clinical and pathological studies on microbes associated with the development and treatment of ASD have yielded inconsistent findings. To our knowledge, there has been no systematic review on this topic. The aim of the current study is to conduct a systematic review to evaluate and summarize findings from studies on the characteristics of the GI microbiome in children with ASD.
2. Methods
2.1 Data retrieval strategies
The process of identifying articles for inclusion in this review is shown in Figure 1. The following key words(in both English and Chinese) were used to search PubMed (1966-2013), Embase (1947-2013), PsycINFO(1966-2013), ISI web of knowledge (1994-2013), Ovid/MEDLINE (1970-2013), the Cochrane Library (1967-2013), the Chinese National Knowledge Infrastructure(1979-2013) database, the Chongqing VIP database for Chinese Technical Periodicals (1989-2013), WANFANG DATA (1990-2013), and the China BioMedical Literature Services System (SinoMed) (1978-2013): autism(including ‘Child Development Disorders, Pervasive’,‘autis*’, ‘pervasive development* disorders*’, ‘child schizophrenia’, ‘kanner*’, ‘Rett*’, ‘Asperger*’, ‘zi bi zheng’[an older term for autism in Chinese]), ‘bacteria’,‘bacter*’, ‘microbiology’, ‘microbio*’, ‘microbiome’,‘microbiome*’, ‘microbial’, ‘microorganism*’,‘metagenomics’, ‘metagenome’, ‘metagenom*’. All articles published before October 5, 2013 were included in the search.
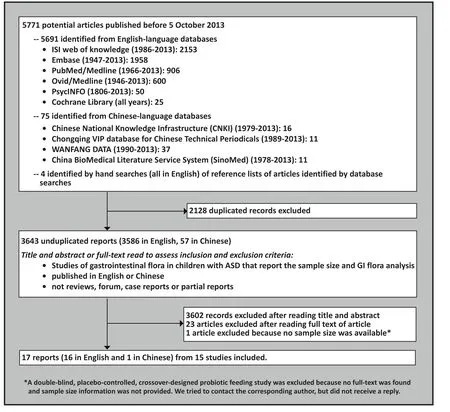
Figure 1. Identification of studies included in the analysis
Identified articles were imported to Endnote X6 to remove duplicated records. Two authors (XC and CL) independently screened titles and abstracts of the remaining articles for potential inclusion. The full text of the articles were downloaded for further assessment when reviewers thought that the article met inclusion criteria or when reviewers were unsure about whether or not the article met the inclusion criteria. The references of all full-text articles were hand searched to identify other potential articles. These two authors then independently read the full-text articles and evaluated them according to the pre-defined inclusion or exclusion criteria listed below. These authors had disagreements about whether or not to include four articles; these disagreements were resolved through discussion.
2.2 Inclusion and exclusion criteria
All included studies: (a) were about the GI bacteria in children diagnosed with ASD; (b) had a group of children diagnosed with ASD, regardless of which diagnostic criteria were used (including but not limited to the Diagnostic and Statistical Manual of Mental Disorders[DSM],[2]the International Statistical Classification of Diseases and Related Health Problems [ICD],[3]and Chinese Classification and Diagnostic Criteria of Mental Disorders [CCMD][4]); (c) had information about sample size and the prevalence of the specific bacteria assessed;(d) were written in English or Chinese. Studies on nonhuman subjects, reviews, case reports, and duplicate publications were excluded.
2.3 Data extraction and evaluation
Two authors (CX and LC) independently extracted data from identified studies including the name of the first author, year of publication, location of the study,sample size, diagnosis of ASD and criteria used to make the diagnosis, sample characteristics (GI symptoms,diet and medication use), source of bacterial samples,testing methods to identify specific bacteria, and the prevalence of different bacteria in children with and without ASD.
The evaluation of the quality of the articles was conducted by the same two authors following the guidelines listed in the article entitled ‘Strengthening the Reporting of Observational Studies in Epidemiology(STROBE)’.[10]For each included study, a total of 22 items were rated based on the content of different parts of the article reporting the study, including the title,abstract, introduction, methods, results, discussion, and other information. Each item counts for one point so the total quality score ranges from 0 to 22. The interrater reliability of the total quality score for the included studies of the two authors was high (ICC=0.90).
2.4 Statistical Analysis
The data were entered into a database and analyzed using Review Manager 5.2.6 software. Considering the discrepancies in methodology across studies,standardized mean difference (SMD) were calculated using different estimation formulas for reports with different sample sizes according to the suggestion of Hozo and colleagues.[11]Studies were considered homogeneous when I2(i.e., the effect size variation attributable to heterogeneity) was less than 50% and the associated p>0.10; in this case a fixed effect model was used to generate pooled estimates. If I2was >50%or p<0.10 the studies were considered heterogeneous so a random effect model was used.[12]No analysis was performed and no pooled estimates were generated when there were less than three studies with relevant data for a particular issue or when the I2was greater than 75%.[13]If there was significant heterogeneity across studies, sensitivity analysis and subgroup analysis was performed to explore the source of heterogeneity if there were a sufficient number of studies in the subgroups. When there were more than 10 studies, a funnel plot was generated to assess publication bias.
3. Results
3.1 General characteristics of included studies
As shown in Figure 1, a total of 3643 unduplicated reports were retrieved. After screening according to the pre-defined inclusion and exclusion criteria, 17 reports were included in the final analysis. Among the 17 studies, the single study from China[14]was an abstract of a master’s thesis; this author was unable to provide either a full text article or any data because the relevant papers had not yet been published; the cases reported(n=6) were, nevertheless, included in our main analysis.Two reports by Williams and colleagues published in 2011[15]and 2012[16]were based on the same sample,and two reports by Wang and colleagues published in 2010[17]and 2011[18]were also based on the same sample. Therefore, data from 15 separate studies were analyzed. All 15 were cross-sectional studies; among them, one[19](n=4) was designed as an open clinical trial,but the GI microbiome assessment was only reported at baseline.
The characteristics and main findings of the studies are shown in Tables 1 and 2. The 15 identified studies were published between 1999 and 2013 in five countries:eight in the United States, three in Australia, two in Poland, one in Britain and one in China. One of the studies had no control group,[20]two studies[19,21]used previously reported data of adults as historical controls,one study[22]had siblings of the ASD children (SIB) as a control group, eight studies[14-16,23-25,27,28,30]used unrelated children without ASD as the control group (CON), and three studies[17,18,25,29]had both sibling and unrelated children without ASD as control groups (i.e., SIB and CON). In total, the 15 studies had a cumulative sample of 805 individuals, including 437 children with ASD,94 siblings of children with ASD, 145 concurrent child controls without ASD, and 129 historical adult controls.Only four studies reported the diagnostic criteria used to determine ASD: three used the DSM criteria[15,18,19]and one used the ICD criteria.[23]
Quantitative analysis on GI bacteria was carried out in 13 studies[14-19,21,22,24-30]; the other two studies[20,23]only described the number and types of cultured bacteria and their colony morphology. Due to the large variation across studies in evaluation methods,sources of specimens, and statistical analyses, it was not appropriate to pool the main results from the studies.
3.1.1 Gender composition of the sample
Among the 15 studies, four studies[14,19,26,30]did not report the gender of the participants, in one study[15,16]participants were all males, and one study[27]only reported the gender of the children with ASD but not that of the control subjects. In the studies that reported the gender of all participants, the range in the malefemale ratio in the three different groups of participants were as follows: in the ASD group the male-female ratio ranged from 2.5:1 to 11:1, in the SIB group it ranged from 0.4:1 to 1.4:1, and in the CON group it ranged from 0.8:1 to 5.7:1.
3.1.2 Characteristics of the combined sample
Table 2 summarizes the clinical and dietary characteristics of participants in the studies for which this information was available. Ten (66.7%) of the 15 studies[15,18,19,22,24-29]reported GI symptoms, but three of them[19,26,27]only reported GI symptoms in the ASD group. Ten (66.7%)studies[18,19,21,22,24-29]reported the use of antibiotic or antifungal medication. Nine (60.0%) studies[15,18,21,22,25-29]reported the dietary restrictions of the children and six (40%) studies[18,22,24,25,28,29]reported on dietary supplements (e.g., probiotics). Only one (6.7%) study[15]reported on participants’ allergic diseases.
3.1.3 Source of specimens
Twelve of the 15 studies (80%) only tested fecal specimens, one study[15,16]did ileal and cecal biopsies,one study[26]examined digestive fluids (gastric, duodenal and empty intestinal fluids), and one study[27]collected both biopsy specimens and fluids.
3.1.4 Methods of assessing GI microorganisms
Among the included studies, seven[19-21,23,24,26,27]evaluated the microorganisms using cultures. Nine studies[14-18,22,25,26,28-30]used sequencing techniques(one of these studies[26]cultured the specimens before sequencing), including fluorescence in situ hybridization(FISH), quantitative real-time polymerase chain reaction(qPCR), high throughput SOLiD sequencing, and bacterial tag encoded FLX amplicon pyrosequencing(bTEFAP).
3.2 Evaluation of study quality and assessment of publication bias
The quality of the reports of the studies was evaluated using the STROBE criteria.[10]The study conducted by Zhang[14]could not be assessed because only the abstract was available. The remaining 14 studies had scores ranging from 9 to 18 (maximum score of 22), with a mean (sd) score of 12.7 (2.7). No study was considered of high quality. The most prominent problems included inadequate or absent explanation of: (a) determination of sample size; (b) sources of the sample and method of selecting the sample (only reported in three studies);(c) timing of the study (including duration of illness,persistence of GI symptoms, concurrent treatment, etc.)and; (d) potential biases (only reported in three studies).The different studies used very different indicators of patients’ GI microbiome so it was not possible to generate a funnel plot of the results.
3.3 GI bacterial characteristics of children with ASD
As shown in Table 1, there were 11 studies[14-19,23-26,28-30]with a combined sample of 562 individuals that found significant differences in GI microorganisms between children with ASD and controls, 3 studies[21,22,27]with a combined sample of 215 individuals that found no differences between children with ASD and controls,and one study[20]with 28 participants that did not have a control group.
3.3.1 Three major bacterial phyla
Four studies reported GI tract bacteria at the phylum level of classification.[15,22,25,28]However, the studies described the results using different statistics: means without standard deviations,[22]means with standard deviations,[25]box graphs,[15]and medians with interquartile ranges.[28]Unfortunately, the relatively large study by Gondalia and colleagues in 2012[22](n=104)only reported means without standard deviations and they were unable to provide us with the original data, so the results from this study could not be combined with those of the other three studies.
Figures 2 to 4 show the results of the ASD and CON groups for the three main phyla – Firmicutes,Bacteroides, and Proteobacteria – using the combined sample of 120 individuals in the three studies. For each study the results of the two groups are compared using standardized mean difference (SMD). However,for all three phyla the results of the three studies were quite different, the main measure of heterogeneity,I2, was 87% for the three results for Firmicutes, 91%for the three results for Bacteroides, and 91% for the three results for Proteobacteria. Given the high level of heterogeneity (I2>75%) and the small number of eligible studies (three) it was not feasible to pool the results in a meta-analysis.
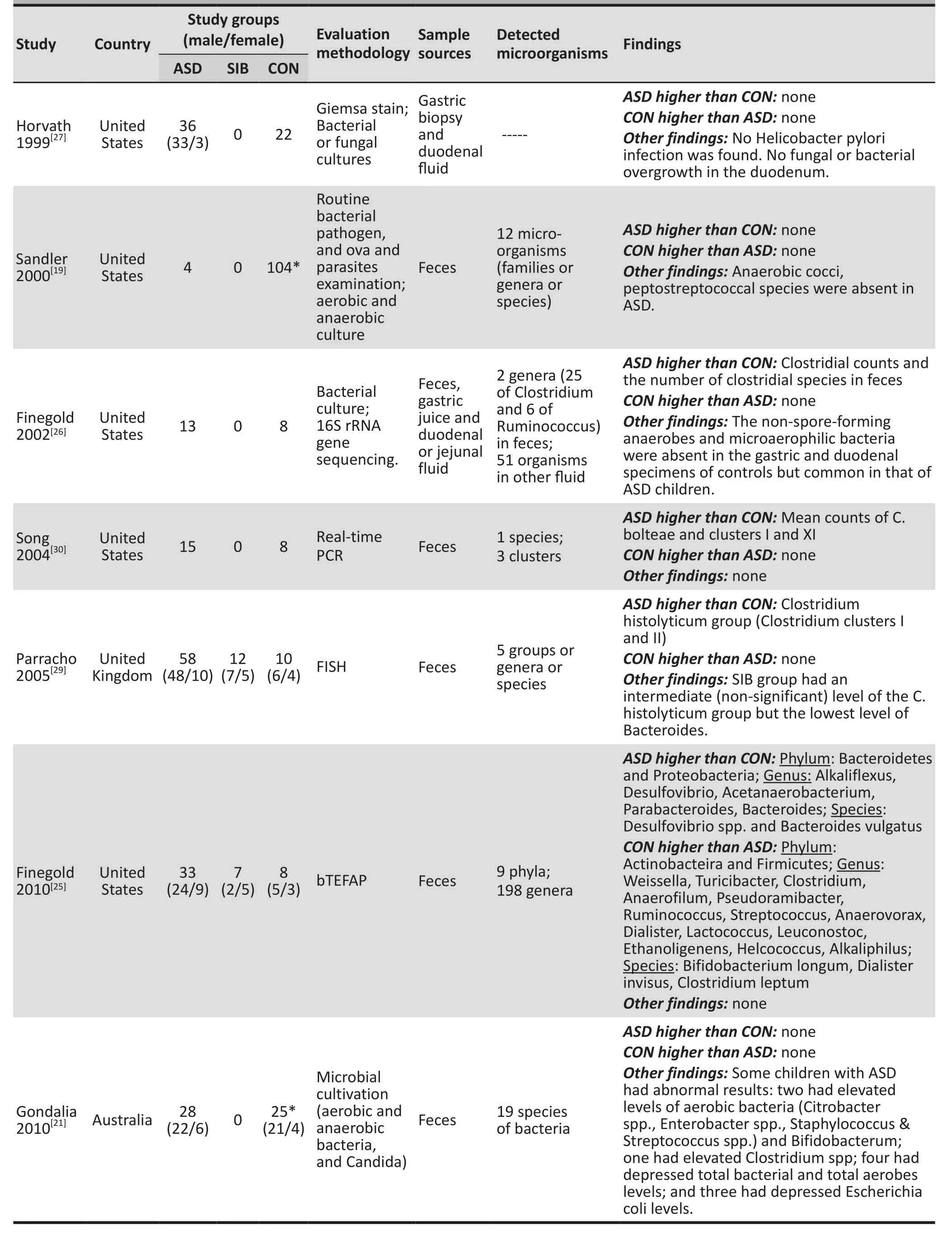
Table 1. Basic characteristics and main findings of the 15 studies included in the systematic review
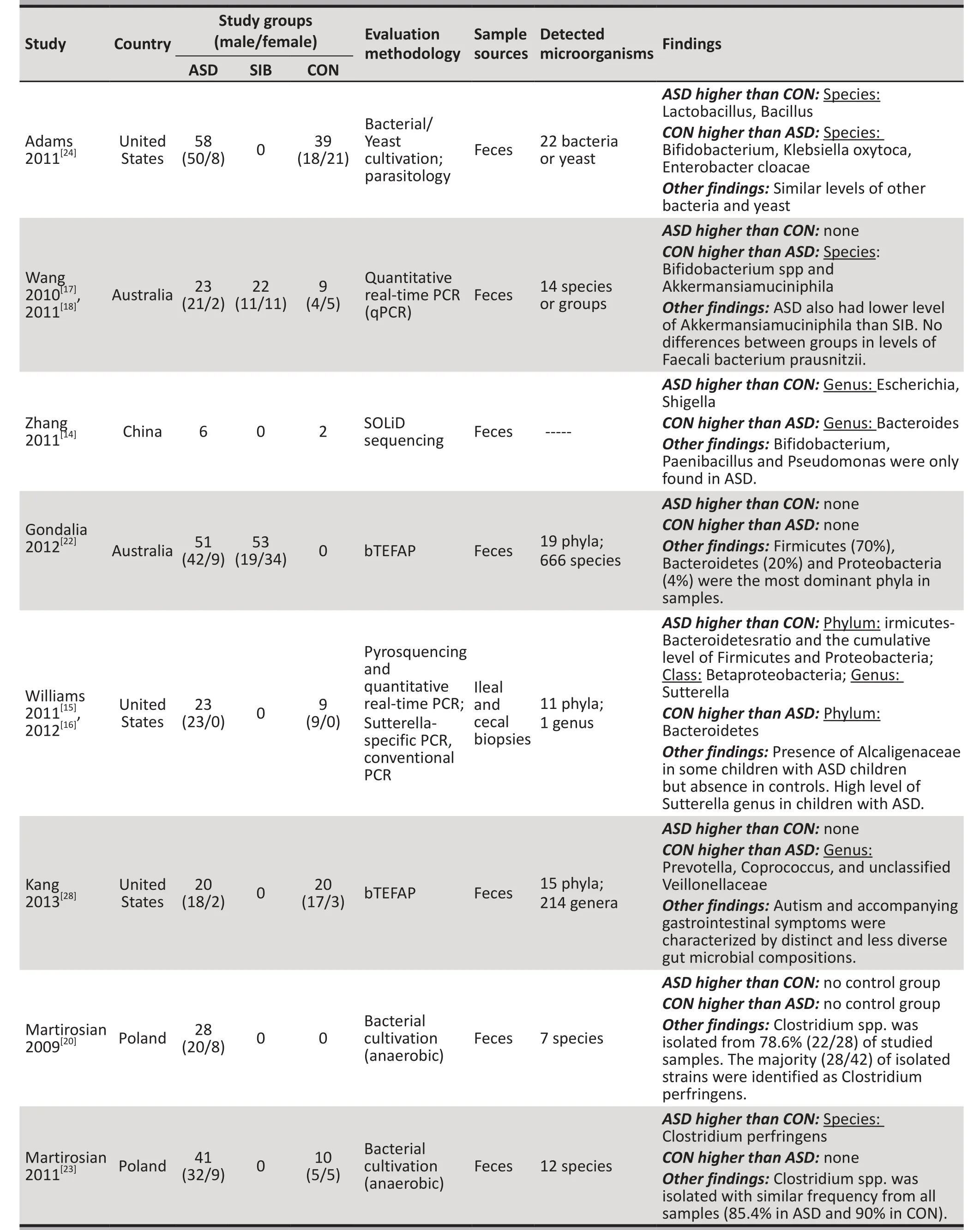
Table 1. Basic characteristics and main findings of the 15 studies included in the systematic review (continued)
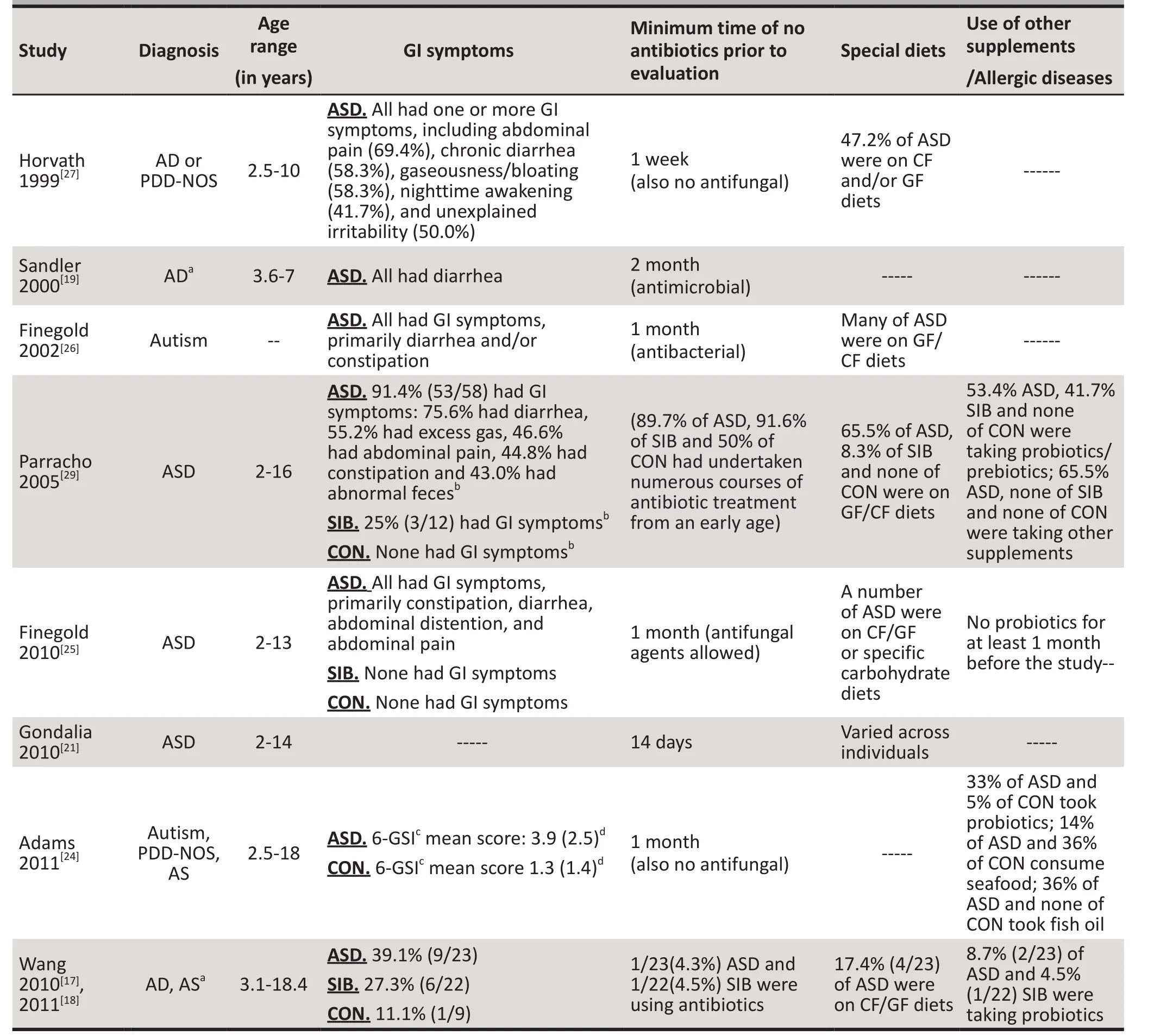
Table 2. Supplemental information provided for 11 of the 15 studies in the systematic review
3.3.2 Clostridium
Although different assessment methods were employed(i.e. culture, real-time PCR or FISH), three studies[26,29,30]with a combined sample of 124 individuals, found higher numbers of some clusters or groups of Clostridium genus bacteria in the stool samples of children with ASD compared to controls. However, a subsequent study conducted by Finegold and colleagues[25](n=48)using bTEFAP yielded the opposite result: compared to controls, a smaller proportion of Clostridium genus and a lower level of Firmicutes phylum (to which Clostridium belongs) were found in children with ASD.
3.3.3 Bifidobacteria
Three studies[17,18,24,25]with a combined sample of 199 individuals reported that the level of the probiotic Bifidobacterium (genus or species) was lower in children with ASD than in controls. But the study by Gondalia and colleagues[21](n=53) found no statistically significant difference in fecal Bifidobacteria levels between children with ASD and a historical control sample of adults without ASD.
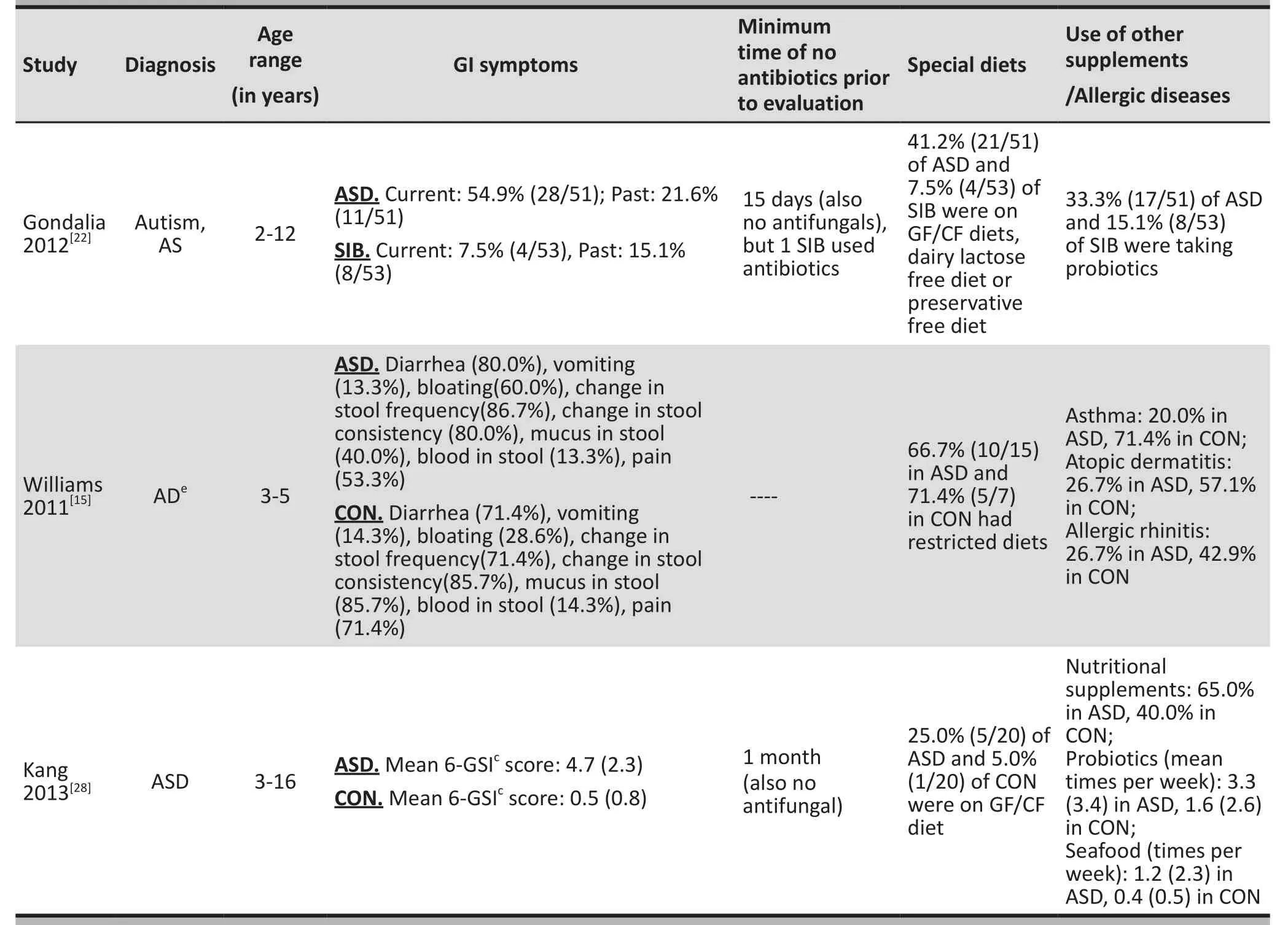
Table 2. Supplemental information provided for 11 of the 15 studies in the systematic review (continued)
3.3.4 Other bacteria
As shown in Table 1, single studies also identified other abnormalities in the GI microbiome of children with ASD: the lack of the anaerobic cocci peptostreptococcus species,[19]and the presence of non-spore-forming anaerobes,[26]microaerophilic bacteria,[26]Paenibacillus,[14]Pseudomonas,[14]and Alcaligenaceae.[16]
3.4 Association between the GI symptoms and the severity of autism
Two studies[24,28]examined the association between GI symptoms and the severity of autism. Both studies used the Autism Treatment Evaluation Checklist (ATEC)to measure the severity of autism and assessed the severity of GI symptoms using the Gastrointestinal Severity Index (6-GSI)[30](which assesses constipation,diarrhea, stool consistency, stool smell, flatulence and abdominal pain). The study by Kang and colleagues[28]also assessed the severity of autism using the Autism Diagnostics Interview-Revised (ADI-Revised), Autism Diagnostics Observation Schedule (ADOS), and Pervasive Developmental Disorder Behavior Inventory (PDD-BI).The study by Adams and colleagues,[24]which included 58 children with ASD, found a strong correlation between the 6-GSI score and the ATEC score (Pearson’s correlation coefficient [r]=0.60, p<0.001). However,the study by Kang and colleagues[28], which included 20 children with ASD, did not find a significant correlation between the 6-GSI score and any of the four measures of autism severity (Spearman’s correlation coefficient [rs]ranged from 0.09 to 0.28; all p>0.05).

Figure 2. Comparisons of the relative abundance of Firmicutes in children with or without Autism Spectrum Disorders (ASD v. CON)
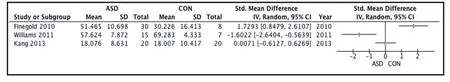
Figure 3. Comparisons of the relative abundance of Bacteroidetes in children with or without Autism Spectrum Disorders (ASD v. CON)
4. Discussions
4.1 Main findings
The characteristics of the GI microbiome in children with ASD is an under-studied field,[5]particularly in China where we were only able to identify the abstract of one small study.[14]The 15 studies that met inclusion criteria for this systematic review varied in the taxonomic level of the assessed organisms (Phylum, Genus, Species) and used different methods to assess the GI microbiome,varying from traditional bacterial cultures to genome sequencing (reflecting the evolution in techniques for assessing GI mircoflora). Methodologically, the studies were generally of poor quality, primarily because of the use of small, unrepresentative samples and little consideration of potential confounders.
Despite the relatively small size of most of the studies (7 of the 15 studies had results for less than 50 individuals), 11 of the 14 studies that had control groups (79%) reported significant differences in the prevalence of different GI microorganisms between children with ASD and controls. However, the variety of assessment methods and differences in the reported organisms made it difficult to pool the results of the different studies. Sufficient information to allow for cross-study comparison was available for 3 studies that used genome sequencing techniques,[15,25,28]but these 3 studies reported conflicting results about the differences in the prevalence of the three main phyla of GI bacteria[32]– Firmicutes, Bacteroidetes and Proteobacteria – between children with ASD and controls. The results about the relative abundance of other GI microorganisms between children with ASD and controls were also inconsistent across the 15 studies. Given the different target organisms assessed,the variety of assessment methods employed, and the heterogeneity of the results for the small number of studies that used similar methods to assess the same class of organisms, it was not possible to conduct a meta-analysis of the results of the studies.
Clearly, in the future more consistency in the methodology across studies will be needed to determine whether or not the GI microbiome of children with ASD is, in fact, significantly different from that of other children. At a minimum studies need to (a) use widely accepted diagnostic criteria for ASD; (b) include a representative sample of ASD children; (c) use validated methods for assessing the severity of ASD and the severity of GI symptoms; (d) select an appropriate control group of children without ASD; (e) use the same sample source (stools) collected in the same manner;(f) report on a common set of organisms (starting with the three main phyla— Firmicutes, Bacteroidetes and Proteobacteria); and (g) collect information on other potential confounding variables including, gender, age,use of antibiotics or antifungal agents, history of allergic and autoimmune conditions, dietary restrictions, and dietary supplements (e.g., probiotics and vitamins). It is certainly possible that an abnormal GI microbiome only occurs in certain subgroups of ASD children or at certain phases in the course of their condition, so there will be need to be sufficiently large samples of subjects included in these studies to identify potentially important subgroups.
4.2 Limitations
The search strategy was quite exhaustive so we expect that there were relatively few relevant studies that were not identified, unless they were published in languages other than English or Chinese. Three high-income countries (i.e., United States, United Kingdom, Australia)accounted for 80% (12/15) of the identified studies and for 89% (718/805) of the combined samples from the identified studies, so it is uncertain whether or not the results would be different if available studies were more internationally representative. The limitations of the results are primarily determined by the limitations in the included studies, which we assessed to be of poor to fair quality (none were good quality). Of the 15 studies included in the review, 7 studies[14,15,20,25,26,28,30]had fewer than 50 subjects (including children with ASD and controls), 1 study did not have a control group,[20]2 studies[19,21]used historical control groups of adults, 1 study was only available as an abstract,[14]and 1 study[22]only provided mean prevalence figures of the different microorganisms without standard deviations (so it could not be compared to other results). As stated previously,the small size, different methods, and heterogeneity of results made it impossible to combine the results of different studies in a meta-analysis.
4.3 Significance
The high reported prevalence of GI symptoms in children with ASD has focused attention on the potential role of GI microorganisms in the onset and development of ASD. To help elaborate a theoretical model that could suggest potential mechanisms for the proposed connection between GI microorganisms and ASD, a number of studies have compared the GI microbiome in children with ASD to that of control subjects. To the best of our knowledge, this paper reports on the first systematic review of these studies. Despite identifying 15 studies that met our inclusion criteria, the small sample sizes, lack of standardization of methods and generally poor methodological quality of the studies makes it impossible to come to definitive conclusions.The majority of studies identified significant differences between children with ASD and controls, but it remains to be proven whether or not the GI microbiome of children with ASD is significantly different from that of controls and, if it is different, what the difference is. Resolution of this theoretically important question will require larger studies of higher quality that use standardized methods for the selection of subjects and the assessment of the GI microbiome.
Conflict of interest
The authors report no conflict of interest related to this manuscript.
Funding
The study was funded by the Research Leadership Development Plan of Shanghai, China (XBR 2011005).
1. Ashwood P, Wills S, Van de Water J. The immune response in autism: a new frontier for autism research. J Leukoc Biol 2006;80(1): 1-15.
2. American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders, 5thed. Washington: American Psychiatric Association; 2013.
3. World Health Organization. ICD-10: International statistical classification of diseases and related health problems. World Health Organization; 2004.
4. Psychiatry Branch of Chinese Medical Association. Chinese Classification and Diagnostic Criteria of Mental Disorder, 3thed. Jinan: Shandong Science and Technology Press, 2001. (In Chinese)
5. Chaidez V, Hansen RL, Hertz-Picciotto I. Gastrointestinal problems in children with autism, developmental delays or typical development. J Autism Dev Disord, 2013. [Epub 2013 Nov 06]
6. Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. Proc Natl Acad Sci USA 1998;95(12): 6578-6583.
7. Douglas-Escobar M, Elliott E, Neu J. Effect of intestinal microbial ecology on the developing brain. JAMA Pediatr 2013;167(4): 374-379.
8. Cryan JF, Dinan TG. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci 2012;13(10): 701-772.
9. Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, et al. Microbiota modulate behavioral and pysiological abnormalities associated with neurodevelopmental disorders. Cell 2013;155: 1-13.
10. Vandenbroucke JP, Von Elm E, Altman DG, Gøtzsche PC,Mulrow CD, Pocock SJ, et al. Strengthening the Reporting of Observational Studies in Epidemiology (STROBE):explanation and elaboration. PLoS medicine 2007;4(10):e297.
11. Hozo SP, Djulbegovic B, Hozo I. Estimating the mean and variance from the median, range, and the size of a sample.BMC Med Res Methodol 2005;5: 13.
蒋曹清 男,1973年出生于湖南省永州市,博士,现为广西财经学院教授,主要研究方向为形式化方法,程序分析,数据挖掘.
12. Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ 2003;327(7414): 557-560.
13. Higgins JPT. Strategies for addressing heterogeneity. In:Higgins JPT, Green S, editors. Cochrane Handbook for Systematic Reviews of Interventions, version 5.1.0. Oxford:The Cochrane Collaboration; 2011.
14. Zhang RM. A preliminary study of intestinal micromicrobiome and autism etiology. Abstract of Master’s Thesis. Southeast University; 2011. (In Chinese)
15. Williams, BL, Hornig M, Buie T, Bauman ML, Cho Paik M,Wick I,et al. Impaired carbohydrate digestion and transport and mucosal dysbiosis in the intestines of children with autism and gastrointestinal disturbances. PLoS One 2011;6(9): e24585.
16. Williams BL, Hornig M, Parekh T, Ian Lipkin W. Application of novel PCR-based methods for detection, quantitation,and phylogenetic characterization of Sutterella species in intestinal biopsy samples from children with autism and gastrointestinal disturbances. MBio 2012;3(1): 1-11.
17. Wang L, Christophersen C, Sorich M, Gerber C, Angley M,Conlon M. Gut bacterial and fermentation profiles are altered in children with autism. J Gastroen Hepatol 2010;25:A116.
18. Wang L, Christophersen CT, Sorich MJ, Gerber JP, Angley MT, Conlon MA. Low relative abundances of the mucolytic bacterium Akkermansia muciniphila and Bifidobacterium spp. in feces of children with autism. Appl Environ Microbiol 2011;77(18): 6718-6721.
19. Sandler RH, Finegold SM, Bolte ER, Buchanan CP, Maxwell AP, Vaisanen ML, et al. Short-term benefit from oral vancomycin treatment of regressive-onset autism. J Child Neurol, 2000;15(7): 429-435.
20. Martirosian G, Ekiel A, Aptekorz M, Kazek B, Marszal E,Jankowska-Steifer E, et al. Intestinal anaerobic bacteria and autistic mind: Is there some relations? Medical Science Monitor 2009;15(3): LE2-LE3.
21. Gondalia SV, Palombo EA, Knowles SR, Austin DW. Faecal microbiota of individuals with autism spectrum disorder.Developmental Disorders & Autism 2010;6: 24-29.
22. Gondalia SV, Palombo EA, Knowles SR, Cox SB, Meyer D,Austin DW. Molecular characterisation of gastrointestinal microbiota of children with autism (with and without gastrointestinal dysfunction) and their neurotypical siblings.Autism Res 2012;5(6): 419-427.
23. Martirosian G, Ekiel A, Aptekorz M, Wiechula B, Kazek B,Jankowska-Steifer E, et al. Fecal lactoferrin and Clostridium spp. in stools of autistic children. Anaerobe 2011;17(1): 43-45.
24. Adams JB, Johansen LJ, Powell LD, Quig D, Rubin RA.Gastrointestinal microbiome and gastrointestinal status in children with autism - comparisons to typical children and correlation with autism severity. BMC Gastroenterology 2011;11(22).
25. Finegold SM, Dowd SE, Gontcharova V, Liu C, Henley KE, Wolcott RD, et al. Pyrosequencing study of fecal micromicrobiome of autistic and control children. Anaerobe 2010;16(4): 444-453.
26. Finegold SM, Molitoris D, Song Y, Liu C, Vaisanen ML, Bolte E,et al. Gastrointestinal micromicrobiome studies in late-onset autism. Clin Infect Dis 2002;35(Suppl 1): S6-S16.
27. Horvath K, Papadimitriou JC, Rabsztyn A, Drachenberg C,Tildon JT. Gastrointestinal abnormalities in children with autistic disorder. Journal of Pediatrics 1999;135(5): 559-563.
28. Kang DW, Park JG, Ilhan ZE, Wallstrom G, Labaer J, Adams JB,et al. Reduced incidence of Prevotella and other fermenters in intestinal micromicrobiome of autistic children. PLoS One 2013;8(7): e68322.
29. Parracho HMRT, Bingham MO, Gibson GR, McCartney AL.Differences between the gut micromicrobiome of children with autistic spectrum disorders and that of healthy children.Journal of Medical Microbiology 2005;54(10): 987-991.
30. Song Y, Liu C, Finegold SM. Real-time PCR quantitation of clostridia in feces of autistic children. Appl Environ Microbiol 2004;70(11): 6459-6465.
31. Schneider CK, Melmed R, Barstow LE, Enriquez FJ, Ranger-Moore J, Ostrem JA. Oral human immunoglobulin for children with autism and gastrointestinal dysfunction: a prospective, open-label study. J Autism Dev Disord 2006;36(8): 1053-1064.
32. Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T,Mende DR, et al. Enterotypes of the human gut microbiome.Nature 2011;473(7346): 174-180.
儿童孤独症谱系障碍胃肠道细菌学研究的系统综述
曹歆轶, 林萍, 蒋平, 李春波*
上海交通大学医学院附属精神卫生中心 上海
背景:有报道指出,孤独症谱系障碍(ASD)患儿的胃肠道(GI)症状发生率较高。然而,有关ASD患儿胃肠道微生物的研究结果不尽一致。目的系统复习相关研究结果,分析ASD患儿各种胃肠道微生物的分布特征。方法:检索 PubMed 、Embase、PsycINFO、ISI web of knowledge、Ovid/Medline、Cochrane Library、中国知识资源总库、中国科技期刊数据库、万方数据检索系统,以及中国生物医学文摘数据库,收集有关ASD患者胃肠道微生物的文献,按照预先制定的纳入及排除标准筛选相关研究。采用Review manger 5.2.6软件进行统计分析。结果最终共纳入15 项小样本横断面研究,其中1项来自于中国。在15项研究中,11项研究(合并样本为562例)报道ASD患儿组与对照组的胃肠道细菌患病率有显著性差异,尤其是厚壁菌,类杆菌和变形菌。但是,由于方法学上较大的异质性以及不同研究结果之间的相互矛盾,我们无法汇集结果进行meta分析。结论目前对ASD患儿胃肠道微生物的研究数量和质量都非常有限。然而,的确有一个“信号”表明ASD患儿的胃肠道微生物和没有ASD的儿童是有显著差异的,因此,继续开展此方面的研究是非常有价值的。为了提高研究的效度,减少研究结果的异质性,将来的研究需要增大样本,标准化方法以及评估相关混杂因素,例如胃肠道症状的严重程度,以及药物,特殊饮食和添加剂的使用情况。
Background:A high prevalence of gastrointestinal (GI) symptoms has been reported in children with Autism Spectrum Disorders (ASD). However, results from studies about the GI mircobiome of such children have been inconsistent.Aim:Integrate the results of studies that examine the distribution of different GI microorganisms in children with ASD.Methods:Studies related to the GI microbiome in children with ASD were identified through PubMed,Embase, PsycINFO, ISI web of knowledge, Ovid/Medline, the Cochrane Library, the Chinese National Knowledge Infrastructure (CNKI) database, the Chongqing VIP database for Chinese Technical Periodicals,WANFANG DATA, and the China BioMedical Literature Service System (SinoMed). Studies were screened for inclusion following pre-defined inclusion and exclusion criteria. Software Review Manager 5.2.6 was used for statistical analysis.Results:A total of 15 cross-sectional studies, all of which had relatively small samples, were included in the final analysis. Only one of the included studies was from China. Among the 15 studies, 11 studies (with a combined sample of 562 individuals) reported significant differences between ASD children and controls in the prevalence of GI bacteria, particularly bacteria in the Firmicutes, Bacteroidetes and Proteobacteria phyla. However, due to the substantial heterogeneity in methodology and the often contradictory results of different studies, it was not possible to pool the results into a meta-analysis.Conclusions:To date, studies on the GI microbiome in children with ASD are limited in quantity and quality.There does, however, appear to be a ‘signal’ suggesting significant differences in the GI microbiome between ASD children and children without ASD, so there would be value in continuing this line of research. To improve validity and decrease the heterogeneity of findings, future studies should enlarge sample sizes,standardize methods and assess relevant confounding variables, such as the severity of GI symptoms and the use of medications, special diets and supplements.
10.3969/j.issn.1002-0829.2013.06.003
Shanghai Mental Health Center, Shanghai Jiao Tong University School of Medicine, Shanghai, China
*correspondence: chunbo_li@163.com
(Received: 2013-11-26; accepted: 2013-12-04)

Xinyi Cao obtained her master’s degree in medicine from Fudan University in 2009. She is currently a resident doctor at the Shanghai Mental Health Center and a doctoral student in psychiatry at the Shanghai Jiao Tong University School of Medicine. Her research interests are plasticity in cognition and the conduct of systematic reviews.
* 通讯作者: chunbo_li@163.com
- 上海精神医学的其它文章
- Can cognitive dissonance methods developed in the West for combatting the ‘thin ideal’ help slow the rapidly increasing prevalence of eating disorders in non-Western cultures?
- Case-control study of allele frequencies of 15 short tandem repeat loci in males with impulsive violent behavior
- Results of the parent-rated Strengths and Difficulties Questionnaire in 22,108 primary school students from 8 provinces of China
- Operationalizing the involuntary treatment regulations of China’s new mental health law
- A case of neuroleptic malignant syndrome induced by perospirone
- Introduction to mediation analysis with structural equation modeling

