四溴代对苯二甲酸及2,2′-联吡啶构筑的铜配位聚合物的合成、晶体结构及热稳定性
刘建锋 刘 艳 吕旭燕 高玲玲 胡拖平
(中北大学理学院,化学实验室,太原 030051)
0 Introduction
1 Experimental
1.1 Synthesis of complex
A H2O solution (5 mL) of H2TBTA (10 mg, 0.02 mmol) and 2,2′-bipy (10 mg, 0.064 mmol) was placed at the bottom of a straight glass tube, upon which a solution of Cu(NO3)2·3H2O (20 mg, 0.08 mmol) in methanol (5 mL) was carefully layed. Upon slow evaporation of the solvents, blue block crystals suitable for X-ray diffraction were yielded after 5 d in 65% yield. IR (KBr pellet, cm-1): 3 398 (s),1 610(vs),1 517(s),1 425(s),1 391(s),1 318(s),1 085(m), 839(m),772(w), 725(m), 559(m). Anal. Calcd. for C18H12CuN2O6Br4(%): C, 29.37; H, 1.63; N, 3.80. Found(%): C,30.11; H, 1.671; N, 4.86.
1.2 Structure determination
Crystallographic data for complex (size: 0.15 mm×0.14 mm×0.10 mm) were collected on a Bruker Smart APEXII CCD diffractometer equipped with a graphite-monothematic Mo Kα radiation (λ=0.071 073 nm) at room temperature. Semi-empirical absorption correction was applied (SADABS) and the program SAINT was used to reduce the data[19]. The structures was solved by direct methods using the SHELXS program of SHELXTL[20]package and refined by fullmatrix least-squares techniques with SHELXL. The metal atoms in the complex were located from E-maps,and other non-hydrogen atoms were located in successive difference Fourier syntheses and refined with anisotropic thermal parameters on F2using SHELX-97[21]. The organic hydrogen atoms were generated geometrically and refined as riding. A summary of the crystallographic data and structure refinement of complex is listed in Table 1. The selected bond lengths and bond angels are listed in Table 2.
CCDC: 870487.

Table 1 Crystal data and structure refinement for complex
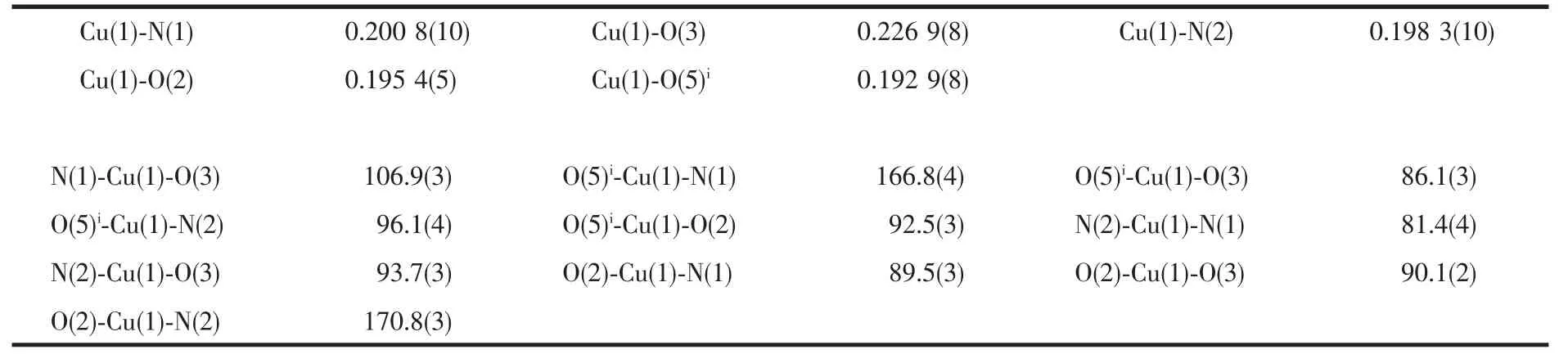
Table 2 Selected bond lengths (nm) and bond angles (°) for complex
2 Results and discussion
2.1 IR spectrum of the complex
In the IR spectrum of complex, the broad peak centered at 3 398 cm-1indicates the O-H characteristic stretching vibrations of uncoordinated water molecular.The asymmetric stretching vibrations(νas(COO)) and its symmetric stretching vibrations (νs(COO)) of H2TBTA appear at 1610 and at 1 391 cm-1, respectively. Their difference Δν(νas(COO-)-νs(COO-)=220 cm-1)between the asymmetric stretching vibrations and the symmetric stretching vibrations shows that carboxylic acid ligand adopts a monodentate mode. The very characteristic bands ν(C=N), ν(C=C) and δ(C-H) of 2,2′-bipy ligand moved from 1 419,1 457 and 758 cm-1to 1 517,1 425 and 772 cm-1indicate 2,2′-bipy ligand coordinated with Cu(Ⅱ)ion[22].
2.2 Crystal structure of the complex

Fig.1 Asymmetric unit of complex
Complex crystallizes in orthorhombic system,Pna21space group. The scheme of the asymmetric unit of complex is in Fig.1. The asymmetric unit of complex consists of one Cu(Ⅱ)ion, one TBTA2-anion,one 2,2′ -bipy molecule, one coordinated and one uncoordinated water molecules. The central Cu(Ⅱ)ion is five-coordinated by two oxygen atoms from two coordinated TBTA2-anions, respectively, two nitrogen atoms from one 2,2′-bipy molecule and one oxygen atom from coordinated water molecule. The carboxylate groups of H2TBTA ligand are deprotonated and coordinate by the monodentate mode. The adjacent Cu (Ⅱ)centers are connected by the TBTA2-anions bridge to afford a 1D chain motif (Cu…Cu=0.822 4 nm) (Fig.2).

Fig.2 1D polymeric chain in complex
In the 1D chain, one pyridine of 2,2′-bipy is parallel to the plane of benzene ring of TBTA2-.Moreover, the 1D chain is stabilized through weak π…π interactions between benzene ring of TBTA2-and 2,2′-bipy. The distance of center to center, Cg(1) …Cg(2), is 0.391 4(5) nm and the dihedral angle is 8.9370° (Cg(1) is the ring C(12)-C(13)-C(14)-C(15)-C(17)-C(18) from TBTA2-, Cg(2)is the ring N(2)-C(1)-C(2)-C(3)-C(4)-C(5) of 2,2′-bipy). The perpendicular distance of Cg(1) on Cg(2) is 0.3512 6(3) nm (Fig.3).
The H-bonding interactions between uncoordinated water molecule and coordinated water molecule and between uncoordinated water molecule and H2TBTA ligand firmly fix the uncoordinated water molecule on 1D chain. In addition, the H-bonding interactions between the hydrogen atoms of 2,2′-bipy and the oxygen atoms of coordinated water and H2TBTA molecules further stabilize the 1D chain (Fig.3). The adjacent 1D arrays are further extended to 2D supramolecular framework by weak H-bonding interactions (O(1)-H(1A)…O(6)v,v-1/2+x, -3/2-y, z;O(2)-H(2B)…O(4)vi,vi-1/2+x, -1/2-y, z). The data of all H-bonding interactions are listed in Table 3.

Table 3 Hydrogen bond lengths and bond angles for complex
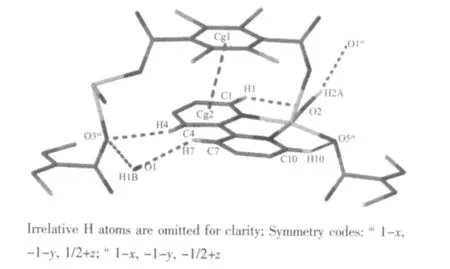
Fig.3 π-π interaction and H-bonding interactions of complex
2.3 Thermogravimetric analysis
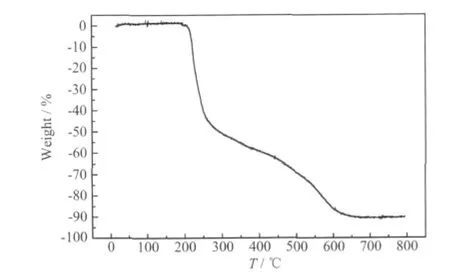
Fig.4 Thermogravimetric analyses for complex
Thermogravimetric analysis has been measured for complex (Fig.4). The complex is stable up to 203℃, The weight loss of 70.06% from 203 to 508 ℃corresponds to the loss of coordinated TBTA2-,coordinated and uncoordinated water molecules(Calcd. 70.12%). The weight loss of 20.27% from 508 to 657 ℃dues to the removal of coordinated 2,2′-bipy molecules (Calcd.21.24%).No weight loss is observed after 657 ℃, and the resulting residue is copper oxide(CuO) (Obsd. 9.68 %, Calcd. 10.82%).
2.4 Powder X-ray diffraction
The results of powder X-ray diffraction (PXRD)analysis of the complex are shown in Fig.5. The PXRD analysis confirms the product to be singlephase. The stimulated PXRD pattern based on the crystal structure analysis allow identification via a comparison of the experimental and computed powder diffraction patterns.
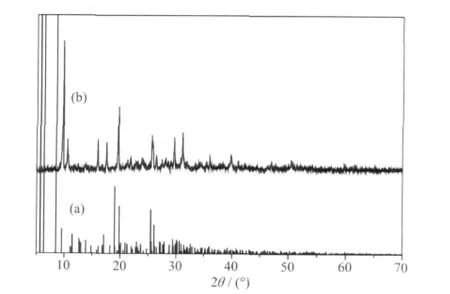
Fig.5 PXRD patterns for complex (a) simulated PXRD pattern; (b) experimental PXRD pattern
[1] Moulton B, Zaworotko M J. Chem. Rev., 2001,101:1629-1658
[2] Kitagawa S, Kitaura R, Noro S I. Angew. Chem., Int. Ed.Engl., 2004,43:2334-2375
[3] Evans O R, Lin W. Acc. Chem. Res., 2002,35:511-522
[4] Spencer E C,Howard J A K,Yaghi O M,et al.Chem.Commun.,2006:278-280
[5] Eddaoudi M, Kim J, Rosi N, et al. Science, 2002,295:469-472
[6] Bourne S A, Lu J J, Mondal A, et al. Angew. Chem., Int. Ed.Engl., 2001,40:2111-2117
[7] Vodak D T, Braun M E, Kim J, et al. Chem. Commun., 2001:2534-2535
[8] Chen S C, Zhang Z H, Huang K L, et al. Cryst. Growth Des.,2008,8:3437-3445
[9] Li C P, Tian Y L, Guo Y M. Inorg. Chem. Commun., 2008,11:1405-1408
[10]Li C P, Tian Y L, Guo Y M. Polyhedron, 2009,28:505-510
[11]Wang X L, Bi Y F, Lin H Y, et al. Cryst. Growth Des.,2007,7:1086-1091
[12]Pan L, Liu H M, Kely S P, et al. Chem. Commun., 2003:854-855
[13]Wang X L, Qin C, Wang E B, et al. Angew. Chem., Int. Ed.Engl., 2005,44:5824-5827
[14]Gomez-Lor B, Gutierrez-Puebla E, Iglesias M, et al. Chem.Mater., 2005,17:2568-2573
[15]Rafael A A, Zhu S R, Kevin K K, et al. Inorg. Chem.Commun., 2007,10(12):1527-1530
[16]Lin Z Z, Chen L, Jiang F L, et al. Inorg. Chem. Commun.,2005,8:199-201
[17]Wang Y Q, Cao R, Sun D F, et al. J. Mol. Struct., 2003,657:301-309
[18]Li X J, Sun D F, Cao R, et al. Inorg. Chem. Commun.,2003,6:908-911
[19]Sheldrick G M. SADABS 2.05, University of Göttingen,Germany, 1996.
[20]SHELXTL 6.10,Bruker Analytical Instrumentation,Madison,Wisconsin, USA, 2000.
[21]Sheldrick G M. SHELXL97, Program for the Refinement of Crystal Structure, University of Gottingen, Germany, 1997.
[22]SHI Zhi-Qiang(石智强), JI Ning-Ning(季宁宁), HE Guo-Fang(何 国 芳), et al. Chinese J. Inorg. Chem.(Wuji Huaxue Xuebao), 2011,27(8):1507-1512

