High-Level Expression and Characteristics of A Thermostable Phytase Mutant from Escherichia Coli K12in Pichia Pastoris
ZHOU Yu-ling,ZOU You,FU Ling,JIANG Wei,ZHAN Fei-xiang,KANG Li-xin,MA Xiang-dong,MA Li-xin
(Hubei Key Laboratory of Industrial Biotechnology,College of Life Sciences,Hubei University,Wuhan 430062,China)
Phytase is an important enzyme used as an additive in animal feed.The enzyme catalyses the sequential hydrolysis of phytate(myo-Inositol 1,2,3,4,5,6-hexakisphosphate,IP6)present in plant material to less phosphorylated myo-inositol derivatives with concomitant release of inorganic phosphate[1].The phosphate is made available to monogastric animals,such as pigs,chickens and fishes,which do not produce phytase and also their microflora cannot degrade phytate.This eliminates the need for external addition of phosphorous to the feed,which incurs costs and also contributes to environmental pollution.
Many phytases from animals,plants and microbes had been isolated and employed as feed additives.In order to obtain phytases with high activity or thermostability,phytase genes from Escherichia coli[2],Bacillus sp.[3],Aspergillus niger[4]and so on were cloned and expressed.Among these phytase genes,the E.coli phytase gene(AppA)had been re-ported to demonstrate the greatest specific activity compared with those from other microorganisms and had been successfully expressed in E.coli[5],Pichia pastoris[6,7]and Streptomyces lividans[2].However,a phytase mutant derived from E.coli K12containing eight amino acid substitutes,for which the activity and thermostability increased significantly compared with the wild type,was of little concerned[8].
In order to increase the expression level and find out the properties of this phytase,here,we described the gene synthesis and characteristics of purified,glycosylated phytase,AppA NR,and its expression in P.pastoris.
1 Experiment
1.1 Strains,chemical agents,enzymes,and media
Pichia pastoris GS115and the vector,pMD18-T,were from Invitrogen (USA).pHBM905Awas stored in our laboratory[9,10].Sodium phytate was purchased from the Sigma.Calcium phytate was prepared as previously described[11].BMGY medium,BMMY medium and minimal dextrose(MD)medium were prepared as described in the manual of the multi-copy Pichiaexpression kit(Invitrogen).
1.2 Optimization and synthesis of gene AppA NR
Based on the synonymous codon usage in P.pastoris and original sequence of E.coli K12mutant[8]containing eight amino acid substitutes,W68E,Q84W,A95P,K97C,S168E,R181Y,N226C,Y277D,the gene was first deduced to its amino acids sequence by GENETOOL and subsequently submitted to oligonucleotide designed with DNAWORKS (http://helixweb.nih.gov/dnaworks/)[12].40Oligonucleotides(Tab.1)were designed for overlapping PCR.The mutant was named as AppA NR with the length of 1299bp,which was too large to synthesis,so the gene was divided into two fragments:658bp and 641bp according to two-step total gene synthesis method[13],then jointed together by PCR.
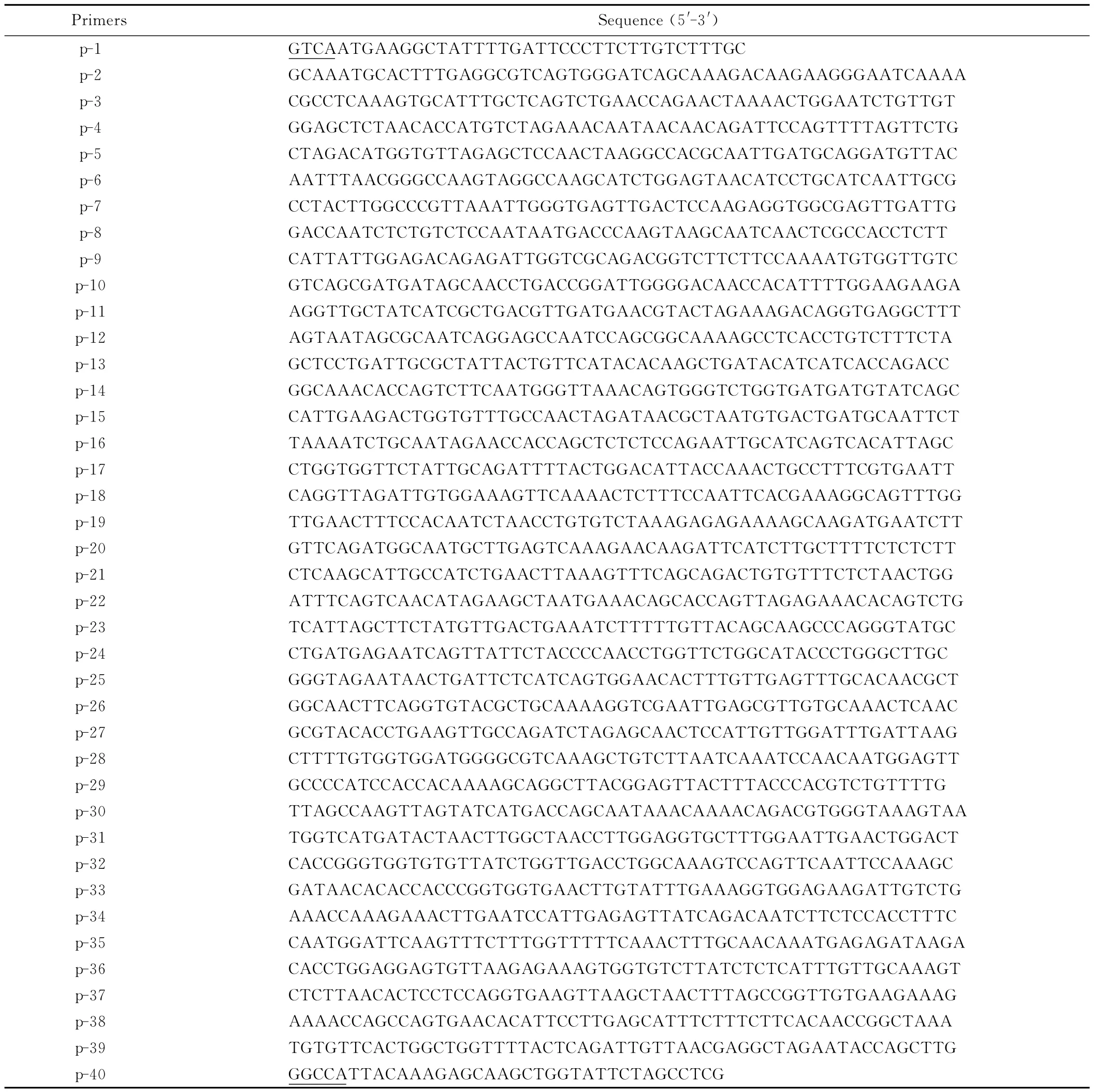
Tab.1 Primers used in this study for the synthesis of gene AppA NR
1.3 Construction of expression plasmid
The amplified primer was designed as follows:AppA-NR-Cpo I (5′-GTCAATGAAAGCGATCT TAATCCCATTTTTATCT-3′),AppA-NR-Not I (5′-GGCCATTACAAACTGCACGCCGGTATGC-3′),which contained CpoI and Not I restriction sites at 5-cohesive ends,respectively.PCR Products were purified and digested with T4DNA polymerase supplemented with 1mmol·L-1dTTP for 20min at 12℃to generate Cpo I or Not I cohesive ends.The expression vector of P.pastoris pHBM905Awith promoter of 5′AOX1,gene was digested by CpoI and Not I restriction enzymes.The treated gene AppA NR was ligated with pHBM905A,downstream of theα-factor secretion signal sequence.This strategy was introduced by Zhang et al.[9].The recombinant plasmid was named as pHBM905A/AppA NR.
1.4 Yeast transformation and gene expression
Transformation of P.pastoris GS115and expression of the recombinant protein were carried out according to the manual of the multi-copy Pichiaexpression kit.Transformants were selected on MD plates without histidine and BMMY plates containing 0.5%calcium phytate by replica plating technique.
1.5 Purification of recombinant phytase
The culture media was centrifuged twice:4000g for 5min and 10 000g for 20min at 4℃.The supernatant was heated in a water bath at 90 ℃for 20s,and then centrifuged at 4 ℃,12 000r· min-1for 10min to remove the thermosensitive protein.The protein was exchanged into a new buffer(100mmol·L-1sodium acetate,pH value of 5.5)using an Amicon Ultra-15centrifugal filter device (20kDa MWCO,Millipore).The molecular weight and purity of phytase were analyzed by 10%SDS-PAGE.The concentration of protein was measured with a protein assay kit from Bio-Rad.
1.6 Glycosylation analysis
Deglycosylation was performed by incubating 2.1g of recombinant phytase with endoglycosidase H(Endo-H)at 37 ℃for 2hwithout the denaturation step.The protein was then analyzed by SDS-PAGE and the residual phytase activity was also tested when treated at 37℃,pH value of 5.5for 10min.A mock-treated sample(the enzyme was incubated with buffer but without addition of deglycosylation enzymes for the same period of time and temperature as the digested sample)was also included.
1.7 Characterization of AppA NR fromP.pastoris
Phytase activity was determined at 37 ℃,pH value of 5.5for 30min following the method as described[14,15].One unit of phytase activity was defined as the amount of enzyme that liberated 1μmol of inorganic phosphate per minute.The relative activity was calculated by comparing the residual phytase activity after each treatment to that of the untreated enzyme.
The optimal temperature was determined by incubating pytase in 0.5% (mass fraction)sodium phytate in 100mmol·L-1sodium acetate with pH value of 5.5for 30min at varying temperatures(25~70℃).
For optimization of pH value,the reaction was performed under 37℃for 30min in 100mmol·L-1of different media with various pH values:HCl(pH value 1.5~2.5),glycine-HCl(pH value 2.5~4.0),sodium acetate(pH value 5.0~6.0)and MOPS(pH value 6.5~8.0).
To investigate the thermostability profile,the 100-fold dilute phytase was heated in water at 95 ℃for 60min in the absence of substrate and the residual phytase activity at different time intervals was measured at 37 ℃.
Protease resistance was tested by incubating AppA NR with pepsin(80mmol·L-1glycine-HCl,pH value of 2.5)or trypsin (80mmol·L-1NH4H2CO4)for 2hat 37℃at a mass ratio of protease to phytase of 1∶100.
The effects of various metal ions and chemical agents on phytase activity were tested by applying appropriate metal salts and chemical agents with the final concentration of 5mmol·L-1,either 3.0%or 0.15%(mass fraction),respectively.The degree of inhibition or activation of enzyme activity was expressed as a percentage of enzyme activity of the control samples(without addition of metal ion or chemical agents).
1.8 Comparative studies on the properties of phytases expressed in Pichia pastoris
The two phytases,AppA and AppA NR,which were both expressed in Pichia pastoris,were compared in thermostability,pH value stability and protease resistance under the same conditions as abovementioned.
2 Results and analysis
2.1 Two-step total gene synthesis of gene AppANR
The phytase gene mutant of E.coli K12is a DNA sequence of 1299bp and encodes a protein of 433amino acids.The coding region was synthesized according to the codon usage bias of P.pastoris,without changing the amino acid sequence.The new gene was referred to as AppA NR from now on(GenBank accession number JX312297).The two fragments with length of 658bp and 641bp were named as AppA NR-1and AppA NR-2,respectively,and synthesized at the first step,and then the fulllength was jointed together by PCR.The electrophoresis and sequence analysis showed that it was correct,and in accordance with the expected.
2.2 Yeast transformation and screen
The phytase gene was cloned into a P.pastoris expression vector pHBM905Aand verified for the correct sequence before being electroporated into P.pastoris GS115.One transformant with the largest halo on the BMMY plate containing 0.5%calcium phytate was chosen for shake-flask fermentation.After 144hof 1% (volume ratio)methanol induction,the yield reached its highest value at 10mg·mL-1with the activity of(980±5.30)U·mL-1in BMMY medium at the flask scale.The culture was centrifuged and the supernatant was electrophoresised on 12%SDS-PAGE.SDS-PAGE analysis indicated that phytase comprised the major protein present in the broth and the molecular weight was about 70kDa(Fig.1a),which was larger than the calculated molecular weight(47.4kDa).One possibility was that the enzyme produced from P.pastoris was glycosylated.Deglycosylation of the recombinant enzyme by Endo-H (removes only high mannose and some hybrid types of N-linked)resulted in a single band with an apparent size of 47kDa(Fig.1b).

Fig.1 SDS-PAGE Analysis
2.3 Characterization of recombinant AppA NR
The effects of temperature and pH value on the activity of the recombinant AppA NR were showed in Fig.2.
The optimal reaction temperature of phytase was 57℃ ,and the enzyme remained above 50%of activi-ty when temperature rised from 37℃to 65℃ (Fig.2a).The enzyme showed good activity at two optimal pH values of 3.5and 6.0,and was inactive below pH value of 1.5or above pH value of 7.5.The activity at optimal pH value of 6.0was 20%larger than that at pH value of 3.5(Fig.2b).
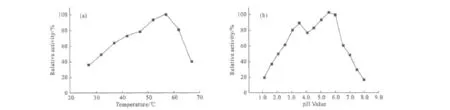
Fig.2 The effects of temperature and pH value on the activity of the recombinant AppA NR phytase
Effects of metal ions and chemical agents on the activity of recombinant AppA NR phytase was showed in Tab.2.
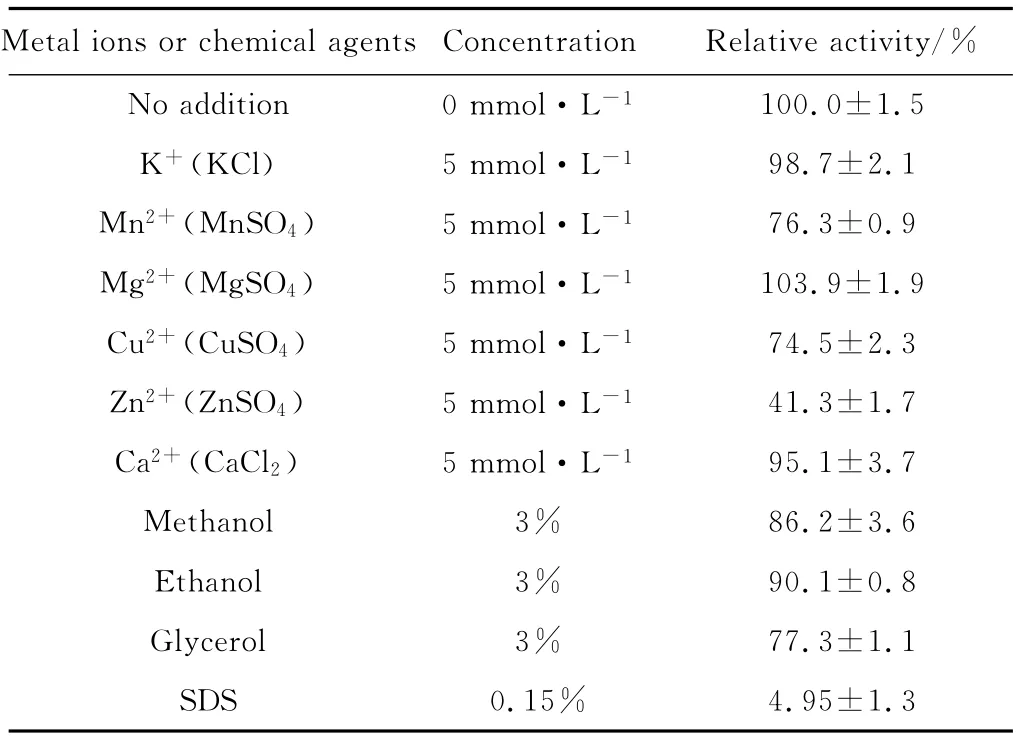
Tab.2 Effects of metal ions and chemical agents on the activity of recombinant AppA NR phytase
K+,Mg2+and Ca2+ions showed little or no inhi-bition effects on enzyme activity and Zn2+inhibited the most inhibition effect,while Mn2+and Cu2+ions decreased enzyme activity by 20%and 30%,respectively (Tab.2).For chemical agents assayed,SDS showed a significant inhibition effect on the phytase activity and almost lowered enzyme activity to zero,while methanol,ethanol and glycerol showed slight inhibition effects on enzyme activity.
2.4 Comparison of the enzymological properties
The comparison of the thermostability and pH value stability of AppA NR and AppA was showed in Fig.3.The thermostability of the recombinant phytases were investigated by incubating the enzyme at 95 ℃for 5min,10min,20min,30min and 60min.The enzyme activity at the zero time-point was defined as 100%.pH Value stability was performed by incubating the enzyme in various buffers.The maximum value was 100%.Values are the means of three independent experiments±standard deviations.

Fig.3 Comparison of the thermostability and pH value stability of AppA NR and AppA
The results of thermostability showed that AppA NR remained more than 85%of its activity,while AppA completely lost the activity after 20min (Fig.3a).The results of pH value stability showed that the two enzymes remained active at a wide pH value range of 1.5~8.0 (Fig.3b).However,low pH value(below pH value of 4.0)showed less effect on AppA than that on AppA NR,while AppA NR remained more activity than that of AppA above pH value of 4.0.
Since phytase is commonly used as feed supplement,proteolytic resistance of AppA and AppA NR were tested during the treating process with protease at a mass ratio of protease to phytase of 0.01∶1for 120min.
As shown in Fig.4a,both of the two phytases showed their resistance to pepsin as their activity were almost unchanged during 120min.On the other hand,the two phytase were sensitive to trypsin,AppA NR and AppA lost 55%and 75%of their activity after incubating with trypsin at 120min,respectively(Fig.4b).
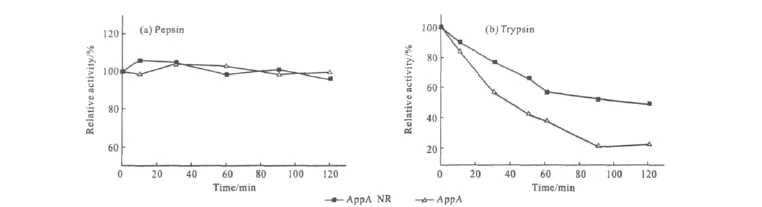
Fig.4 Comparison of the proteases resistance of AppA NR and AppA
2.5 Discussion
A large quantity of the enzyme protein was obtained through expression in P.pastoris,allowing us to obtain adequate pure protein samples in order to analyze enzymatic characteristics.However,it is often difficult for proteins to express highly outside their original context because of codon biases in different hosts.A previous study indicated that the expression level of protein correlated with the operating frequencies of the preferred genetic codons in yeast and codons of low usage could decrease expression level[16].In order to achieve high-level expression and activity,the phytase gene was optimized according to the synonymous codon usage in P.pastoris,which could significantly increase the expression of the recombinant protein.Fortunately,we screened a high-level expression recombinant P.pastoris with the yield of 10mg·mL-1and the activity of 980U·mL-1,which was larger than that of the optimized AppA-mwith 422U·mL-1[17]and the wild AppAgene[6]with the activity of 204U·mL-1expressed in P.pastoris at the flask scale.
Not only did the yield increase dramatically,but also thermostability of AppA NR improved significantly compared with that of AppA expressed in P.pastoris.There were several advantages for thermo-stablity in industrial application because the phytase could sustain the heat denaturation step in the regular feed pelleting process,and could increase the solubility of raw materials,and also could decrease the risk of microbial contamination[18].In our work,we compared the properties of AppA and AppA NR expressed in P.pastoris and found that the properties changed slightly except for thermostability.One explanation for this enhancement of thermostability is due to the glycosylation of the target protein,which is a common post-translational modification in P.pastoris[19].SDS-PAGE indicated that the molecular weight of the recombinant AppA NR was larger than the calculated molecular weight(47.4kDa)(Fig.1).The deglycosylation assay also indicated that AppA NR had been glycosylated.Another explanation was that the eight amino acid substitutes really improved the thermostability,just as explained in the original[8].
3 Conclusion
In summary,we have successfully obtained a high yield(10mg·mL-1)of P.pastoris with high activity(980U· mL-1)and thermotolerance and broad pH value range.All these characteristics suggested that the recombinant AppA NR phytase is a possible source of phytase in feed supplement.
[1]Rao D E,Rao K V,Reddy T P ,et al.Molecular characterization,physicochemical properties,known and potential applications of phytases:An overview[J].Crit Rev Biotechnol,2009,29(2):182-198.
[2]Stahl C H,Wilson D B,Lei X G.Comparison of extracellular Escherichia coli AppA phytases expressed in Streptomyces lividans and Pichia pastoris[J].Biotechnol Lett,2003,25(10):827-831.
[3]Tran T T,Mamo G,Mattiasson B,et al.A thermostable phytase fromBacillus sp.MD2:Cloning,expression and high-level production in Escherichia coli[J].J Ind Microbiol Biotechnol,2010,37(3):279-287.
[4]Promdonkoy P,Tang K,Sornlake W,et al.Expression and characterization of Aspergillus thermostable phytases in Pichia pastoris[J].FEMS Microbiol Lett,2009,290(1):18-24.
[5]Yoon S M,Kim S Y,Li K F,et al.Transgenic microalgae expressing Escherichia coli AppA phytase as feed additive to reduce phytate excretion in the manure of young broiler chicks[J].Appl Microbiol Biotechnol,2011,91(3):553-563.
[6]Chen C C,Wu P H,Huang C T,et al.APichia pastoris fermentation strategy for enhancing the heterologous expression of an Escherichia coli phytase[J].Enzyme and Microbial Technology,2004,35(4):315-320.
[7]Luo H Y,Yao B,Yuan T Z,et al.Overexpression of Escherchia coli phytase with high specific activity[J].Chinese Journal of Biotechnology,2004,20(1):78-84.
[8]Short J M,Gray K A,Barton N R,et al.Phytases,nucleic acids encoding them and methods for making and using them[P].USP 2009 0 062 139,2009-03-05.
[9]Zhang G,Mao L,Zhao Y,et al.Characterization of a thermostable xylanase from an alkaliphilic Bacillus sp.[J].Biotechnol Lett,2010,32(12):1915-1920.
[10]Chen X,Zhai C,Kang L,et al.High-level expression and characterization of a highly thermostable chitosanase from Aspergillus fumigatusin Pichia pastoris[J].Biotechnol Lett,2012,34(4):689-694.
[11]Evans W J,Pierce A G.Calcium-phytate complex formation studies[J].Journal of the American Oil Chemists′Society,1981,58(9):850-854.
[12]Hoover D M,Lubkowski J.DNAWorks:An automated method for designing oligonucleotides for PCR-based gene synthesis[J].Nucleic Acids Res,2002,30(10):e43.
[13]Young L,Dong Q.Two-step total gene synthesis method[J].Nucleic Acids Res,2004,32(7):e59.
[14]Oh B C,Choi W C,Park S,et al.Biochemical properties and substrate specificities of alkaline and histidine acid phytases[J].Appl Microbiol Biotechnol,2004,63(4):362-372.
[15]Fugthong A,Boonyapakron K,Sornlek W,et al.Biochemical characterization and in vitro digestibility assay of Eupenicillium parvum(BCC17694)phytase expressed in Pichia pastoris[J].Protein Expr Purif,2010,70(1):60-67.
[16]Sreekrishna K,Brankamp R G,Kropp K E,et al.Strategies for optimal synthesis and secretion of heterologous proteins in the methylotrophic yeast Pichia pastoris[J].Gene,1997,190(1):55-62.
[17]Huang G R,Tian L C,Huang S P,et al.High expression of codon optimized phytase-encoding gene AppA-Pin Pichia pastoris[J].Journal of Hubei University(Natural Science),2007,29(3):290-293.
[18]Viader-Salvado J M,Gallegos-Lopez J A,Carreon-Trevino J G,et al.Design of thermostable beta-propeller phytases with activity over a broad range of pHs and theirs overproduction by Pichia pastoris[J].Appl Environ Microbiol,2010,76(19):6423-6430.
[19]Lehmann M,Pasamontes L,Lassen S F,et al.The consensus concept for thermostability engineering of proteins[J].Biochim Biophys Acta,2000,1543(2):408-415.

