PTEN and PDCD4 are Bona Fide Targets of microRNA-21 in Human Cholangiocarcinoma△
Chang-zheng Liu,Wei Liu, Yi Zheng, Jin-mei Su, Jing-jing Li, Lan Yu, Xiao-dong He, and Song-sen Chen
1Department of Biochemistry, National Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing 100005, China 2Department of General Surgery, 3Department of Rheumatology, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing, 100730, China
MICRORNAs are small, 20- to 24-nucleotide non-coding RNAs found in diverse organisms.1They play important roles in multiple biological processes such as development, differentiation, and cellular stress response.2,3Recent studies have revealed dysregulation of microRNAs correlates to various diseases including cancer.4,5Cholangiocarcinoma (CCA) is the second most common primary hepatic malignancy. Survival in CCA is dismal, usually measured in months. Moreover, the vast majority of patients are diagnosed at late stages, when surgery is not a viable option.6Reports have demonstrated that deregulation of microRNAs is associated with CCA formation and progression.6Therefore, microRNAs could be potential biomarkers for clinical diagnosis.
Dramatic up-regulation of microRNA-21 has also been reported in multiple cancers and a few targets have been experimentally validated, including phosphatase and ten- sin homolog (PTEN), programmed cell death 4 (PDCD4), reversion-inducing-cysteine-rich protein with kazal motifs (RECK), tropomyosin 1 (TPM1), and tissue inhibitor of metalloproteinases 3 (TIMP3).6-12Moreover, functional changes involved in these targets modulated by microRNA-21 were also found in tumorigenesis. Several questions remained unanswered. For example, are the above mentioned targets directly regulated by microRNA-21 in CCA? What is the correlation between microRNA-21 and its effectors in the same CCA tissue? In this study, the expression levels of microRNA-21 were evaluated in both CCA tissues and QBC939 cells by using real-time PCR or locked nucleic acid in situ hybridization (LNA-ISH) analysis. More importantly, we aimed to validate the direct functional targets of microRNA-21 in CCA tissues and cells by using dual-luciferase reporter gene assay, western blot analysis, and immunohistochemistry analysis.
MATERIALS AND METHODS
Human CCA tissue specimen preparation and cell culture
CCA tissue specimens and normal bile duct tissue specimens adjacent to tumor obtained from 20 CCA patients were collected at surgery in Peking Union Medical College Hospital, immediately frozen and stored in liquid nitrogen until RNA extraction. The pathology of these specimens was confirmed by hematoxylin and eosin staining. The normal bile duct tissues were obtained from 3 donors with bile duct stones. The mean age of 13 male and 7 female CCA patients was 55 years. This study was approved by the Committee for the Conduct of Human Research and patient's informed consent was obtained.
The 293A and CCA cell line, QBC939 were conserved in our lab and grown in DMEM (GIBCO, USA) supplemented with 10% fetal bovine serum (Hyclone, USA) at 37°C in 5% CO2cell culture incubator.
RNA isolation and real-time quantitative PCR
Total RNA was extracted from cells and tissues by using Trizol reagent (Invitrogen, USA). Real-time PCR of microRNA-21 was performed as the protocol supplied by ABI (USA). Real-time quantitative PCR analysis was conducted to detect the mRNA expression of PTEN (Forward: 5'- TGCAGAGTTGCACAATATCCTT-3', Reverse: 5'-GTCATCTT- CACTTAGCCATTGGT-3'), PDCD4 (Forward: 5'-ATGAGCAC- AACTGATGTGGAAA-3', Reverse: 5'-ACAGCTCTAGCAATA- AACTGGC-3') and β-actin (Forward: 5'-CGTACCACTGG- CATCGTGAT-3', Reverse: 5'-GTGTTGGCGTACAGGTCTTTG- 3'). For these analyses, 1 μg of total RNA from the cell lines and tissues was converted to cDNA using SUPERSCRIPTTMⅢ First-Strand Synthesis System (Invitrogen) with the following incubations: 65°C for 5 minutes; 0°C for 1 minute; 50°C for 50 minutes; 85°C for 5 minutes. Following cDNA synthesis, real-time quantitative PCR was performed on the ABI Prism 7500 HT Sequence Detection System (Applied Biosystems, USA) using SYBR®Green PCR Master Mix (Applied Biosystems) with the following cycling conditions: 95°C for 10 minutes (initial denaturation), then 40 cycles of 95°C for 15 seconds, and 60°C for 1 minute. Real-time PCR analysis of mircoRNA-21 expression was performed in normal bile duct tissues, paired CCA tissues, and QBC939 cells. U6snRNA served as loading control. Results represented three independent experiments. Mean±SD were obtained from three independent experiments.
Plasmid construction and luciferase reporter gene assay
pRL-TK plasmid containing a cDNA encoding Renilla luciferase and pGL3-control plasmid containing a cDNA encoding Fly luciferase were obtained from Promega, USA. pRL-TK-21 PER plasmid containing microRNA-21 consensus target sequence served as a positive control. The 3'-untranslated regions (3'-UTRs) sequences of the human PTEN or PDCD4 mRNA were cloned into pRL-TK vector between the Not I and Xba I sites and denoted as PTEN_wt or PDCD4_wt, respectively. Primers for pRL-TK-21 PER, PTEN, and PDCD4 are shown in Table 1. Mutation of the referred mRNA sequence was created using a QuickChange Site-Directed Mutagenesis kit (Stratagene, USA). The mutant plasmids were denoted as PTEN_mut and PDCD4_mut, respectively.
MicroRNA-21 mimic and the negative control, denoted as Ctrl-mimic were obtained from Ambion, USA. Anti-microRNA-21 and the corresponding negative control, scrambled oligonucleotide (here denoted as scramble), were products of Exiqon, Denmark.
One day before transfection, 293A cells were plated in 24-well plates at a concentration of 2×104cells per well. These reporter plasmids, PTEN_wt (0.2 μg/well), PDCD4_wt (0.2 μg/well), PTEN_mut (0.2 μg/well), and PDCD4_mut (0.2 μg/well), and pGL-3 control (0.05 μg/well) were cotransfected with either microRNA-21 mimic (50 nmol/L) or scramble (50 nmol/L), respectively. At 24 hours after transfection, cells were lysed for luciferase activity analysis using dual-Glo TM luciferase assay system (Promega, USA). The luciferase activity was normalized with that in cells treated with scramble. Results represented three independent experiments. Mean±SD were obtained from three independent experiments.
To further investigate the interaction between endogenous microRNA-21 and its targets, QBC939 cells were seeded onto 24-well plates (2×104cells per well) the day before transfection. PTEN_wt (0.2 μg/well), PDCD4_wt (0.2 μg/well), PTEN_mut (0.2 μg/well), and PDCD4_mut (0.2 μg/well) were cotransfected with pGL-3 control (0.05 μg/well) into QBC939 cells, respectively. All transfections were carried out in triplicate using Effectene (Qiagen, Germany). Cell lysates were prepared and luciferase activities were measured using the Dual Luciferase Reporter Assay and dual-Glo TM luciferase assay system (Promega, USA).
Western blot
QBC939 cells were transfected with anti-microRNA-21 or scramble. After 48 hours of transfection, Western blot analysis was performed as previously described12and the membranes were blotted with antibodies for 2 hours at room temperature for PTEN, PDCD4, and GAPDH (rabbit polyclonal antibody to GAPDH from Santa Cruz, USA, used in 1/2000 dilution; rabbit polyclonal antibody to PTEN and PDCD4 from CST, USA, used in 1/1000 dilution). The membranes were incubated with the secondary antibody linked with horseradish peroxidase (HRP) against rabbit (1/5000 dilution; Santa Cruz, USA) for 1 hour. Exposure was performed by using Chemiluminescent Western Blotting Kits (Pierce, USA). The signal was quantified by Kodak Imaging software and the ratio of PTEN or PDCD4 to GAPDH was determined.

Table 1. Sequences of primers used for luciferase reporter gene assay
LNA-ISH analysis
LNA-ISH analysis of microRNA-21 expression was performed as described previously.13Five-micrometer-thin sections of formalin fixed, paraffin-embedded tissues adhered to glass slides were deparaffinized in three con secutive xylene baths for 1 minute each, followed by 1 minute each in serial dilutions of ethanol (100%, 100%, 95%, and 95%), and three changes of diethyl pyrocarbonate-treated water. Slides were then immersed in 0.3% H2O2for 30 minutes at room temperature, washed thrice with diethyl pyrocarbonate-treated water, digested with 400 μg/mL proteinase K (Roche, Switzerland) at 37°C for 15 minutes, washed thrice with diethyl pyrocarbonate-treated water, submerged in 95% ethanol for 1 minute, and air-dried completely. Slides were then hybridized in incubation chambers overnight at 37°C in an oven, using 0.2 μmol/L LNA-modified probes diluted with mRNA ISH solution (DAKO, Denmark). After hybridization, slides were rinsed thrice in 0.5×SSC, washed for 30 minutes at 50°C in 0.5×SSC/0.1% Brij35 (Sigma, USA), and rinsed twice in TBS. An anti-FITC horseradish peroxidase (HRP)-conju- gated antibody (DAKO, Denmark) at 1:100 dilution in TBS/1% bovine serum albumin was applied to the slides for 60 minutes at room temperature, followed by three washes in TBS/0.1% Tween 20 (TBS-T). For amplification of antibody signals, FITC-conjugated phenol (fluorescyl-tyra- mide, DAKO, Denmark) was applied to the slides for 30 minutes at room temperature, followed by three washes in TBS-T. Finally, an anti-FITC antibody conjugated to HRP (DAKO, Denmark) was added to the slides for 30 minutes at room temperature, followed by three washes in TBS-T. The reaction products were visualized using a 50 mg/dL 3, 3′-diaminobenzidine tetrahydrochloride solution containing 0.003% hydrogen peroxide.
Immunohistochemistry
Immunohistochemical staining was performed as follows. Paraffin-embedded tissue was pretreated at 65°C for 2 hours, followed by deparaffinization using standard procedures. Antigen retrieval was performed in antigen retrieval solution before application of primary antibodies (rabbit polyclonal antibody to PTEN and PDCD4 from CST, used in 1/200 dilution) for 1 hour at room temperature. Slides were then incubated for 2 hours at room temperature with the secondary antibody against rabbit (1/500 dilution), conjugated with HRP. HRP activity was detected using the Histostain-plus kit (Invitrogen, USA) according to the manufacturer's instructions. Finally, the sections were counterstained with hematoxylin and mounted.
Statistical analysis
Microsoft Excel software was used for statistical analysis. Student's t test (two-tailed) was performed to compare two groups unless otherwise indicated (χ2test). P<0.05 was considered as significant.
RESULTS
Up-regulation of microRNA-21 expression in CCA tissues and QBC939 cells
Real-time PCR analysis showed the expression of microRNA-21 was consistently higher (about 5.6-fold) in CCA tissues compared with the tumor-adjacent normal tissues (P<0.05, Fig. 1B). Furthermore, microRNA-21 expression in QBC939 cells was also elevated compared with the normal bile duct tissues (P<0.05, Fig. 1C).
PTEN and PDCD4 are direct targets of microRNA-21 in CCA cells
To validate PTEN and PDCD4 as bona fide targets of microRNA-21, we cloned the predicted target regions within PTEN's and PDCD4's 3'-UTR downstream of a Renilla luciferase reporter gene. Reporter gene assay in 293A cells revealed microRNA-21-dependent repression of these 3'-UTRs, and mutation of these microRNA-21 sites abrogated the reduction in luciferase activities (Fig. 2A).
To further verify this finding, we examined the regulation of endogenous microRNA-21 on the binding sites located at 3'-UTR of PTEN and PDCD4 mRNA compared with seed mutants by using dual luciferase reporter gene assay. We observed that endogenous microRNA-21 in QBC939 cells could inhibit the luciferase activity of a reporter gene with the wild-type seed sequence but not for the mutants (Fig. 2B). Taken together, these findings indicated the specificity of the interaction between microRNA-21 and its target regions located at the 3'-UTRs of PTEN and PDCD4 mRNA.
Increased endogenous protein levels of PTEN and PDCD4 in QBC939 cells through inhibiting microRNA-21
QBC939 cells transfected with anti-microRNA-21 achieved a greater than 80% inhibition of microRNA-21 levels (Fig. 3A). Western blot showed the expressions of PTEN and PDCD4 protein in QBC939 cells after anti-microRNA-21 treatment were increased compared with the scramble-treated QBC939 cells (Fig. 3B), while the mRNA expression level of these targets did not show the same trend (Fig. 3C).

Figure 1. Relative ratio of microRNA-21 expression in 20 cholangiocarcinoma (CCA) and matched normal tissue specimens, as well as in QBC939 cells detected by using real-time PCR analysis. A. Relative ratio of microRNA-21 mRNA expression in the CCA tissues is higher than that in the matched normal tissues. B. Average ratio of microRNA-21 mRNA expression in the matched normal tissues and CCA tissues. U6 snRNA serves as control.Data were expressed as mean±SD (n=3), *P<0.05. C. Relative ratio of microRNA-21 mRNA expression in normal bile duct and QBC939 cell line. U6 snRNA serves as control. Data were expressed as mean±SD (n=3), *P<0.05.

Figure 2. microRNA-21 inhibits the activity of PTEN and PDCD4 reporters. A. Ectopic expression of microRNA-21 induced by microRNA-21 mimic markedly suppresses the activity of wild-type PTEN and PDCD4 reporters. Error bars are derived from six experiments in triplicate.*P<0.05, **P<0.01 compared with relative luciferase activity in 293A cells transfected with Ctrl-mimic. B. Endogenous microRNA-21 remarkably inhibits the activity of wild-type PTEN and PDCD reporters. Error bars are derived from six experiments in triplicate.*P<0.05, **P<0.01 compared with relative luciferase activity in QBC939 cells transfected with PTEN_wt or PDCD4_wt.
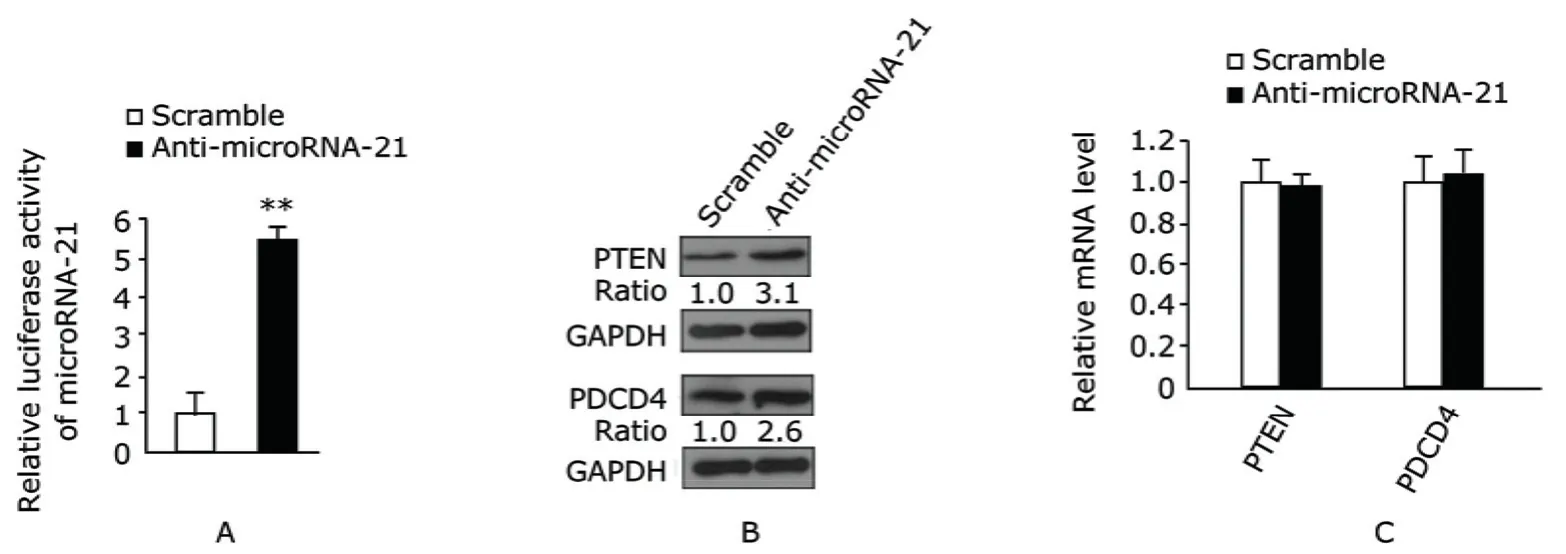
Figure 3. microRNA-21, PTEN, and PDCD4 expression levels in QBC939 cells after transfection of anti-microRNA-21 or scramble detected using Western blot and real-time PCR. A. Relative luciferase activity of microRNA-21 in QBC939 cells after cotransfection of pRL-TK-21PER with anti-microRNA-21 for 48 hours was markedly inhibited. Error bars are derived from six experiments in triplicate. **P<0.01 compared with relative luciferase activity in QBC939 cells transfected with scramble. B. After 48 hours of transfection, Western blot analysis showed PTEN and PDCD4 expressions in QBC939 cells transfected with anti-microRNA-21 markedly increased compared with cells transfected with scramble. C. Relative mRNA expressions of PTEN and PDCD4 in QBC939 cells transfected with anti-microRNA-21 or scramble for 48 hours respectively were analyzed by using real-time PCR. β-actin serves as control. The specificity of every real-time PCR was assessed with melting a curve in a tentative test (n=3).
A negative correlation between microRNA-21 and PTEN, PDCD4 in the same CCA sample
Real-time PCR (Fig. 4A) and LNA-ISH analysis (Fig. 4B) indicated that microRNA-21 expression was markedly up-regulated in CCA sample 1(#1) and sample 4(#4) about 3.5-fold (P<0.05) and 2.9-fold (P<0.01), compared with the matched normal tissues, respectively. In contrast, PTEN and PDCD4 expressions were reduced as compared to the matched normal tissues, shown by immunohistochemistry analysis (Fig. 4C).
DISCUSSION
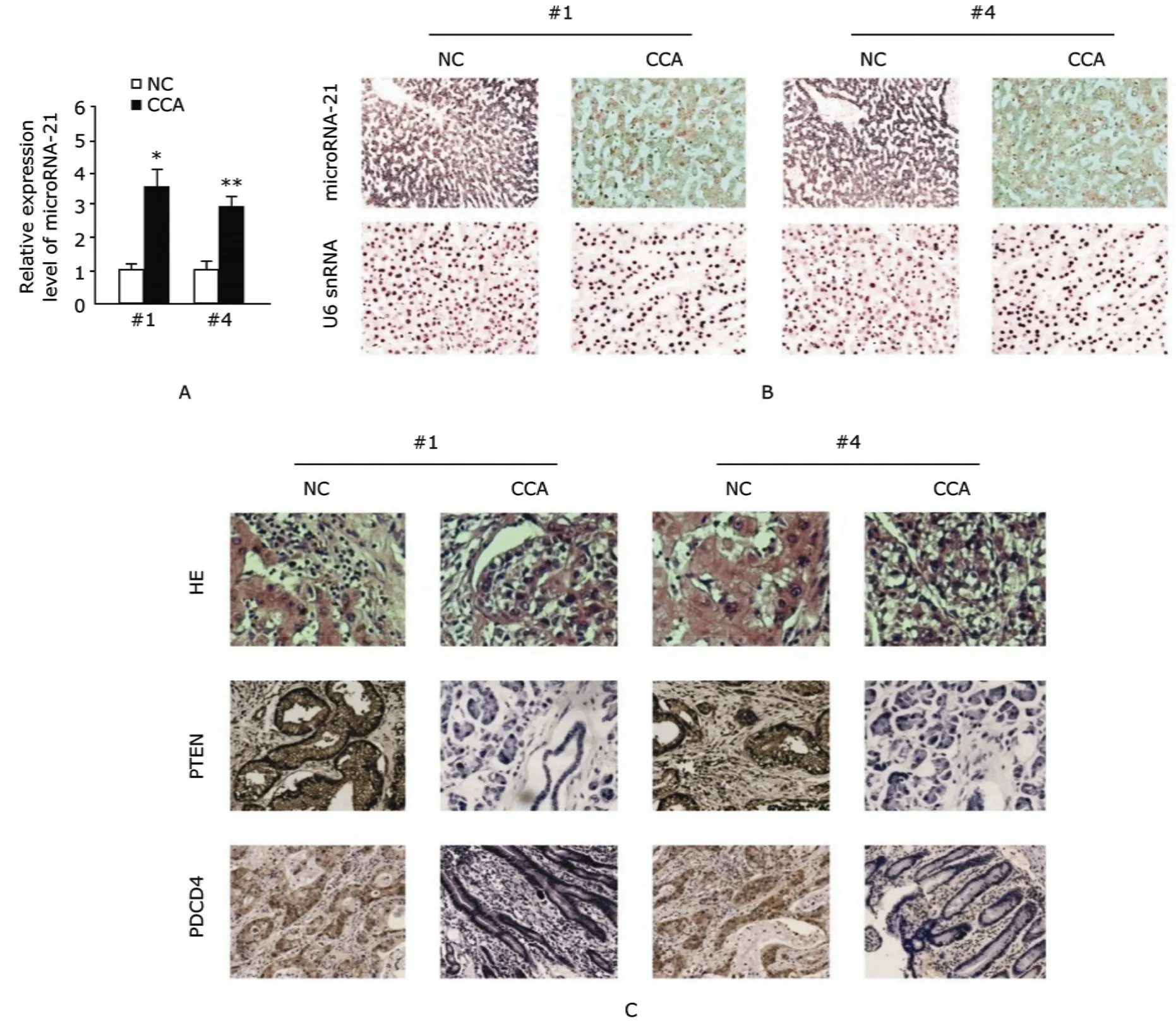
Figure 4. A negative correlation between microRNA-21 and PTEN, PDCD4 is indicated in the same CCA samples. The CCA sample 1 and 4 were denoted as #1 and #4, respectively. NC: matched normal tissue. A. Relative expression level of microRNA-21 in 2 pairs of CCA tissues and matched normal tissues analyzed by using real-time PCR. U6 snRNA serves as control. The specificity of every real-time PCR was assessed with melting a curve in a tentative test (n=3).*P<0.05, **P<0.01 compared with NC. B. LNA-ISH analysis was performed to detect microRNA-21 in 2 pairs of CCA tissues. U6 snRNA serves as control. HRP ×20 C. Expression of PTEN and PDCD4 was studied by using immunohistochemistry, which showed that PTEN and PDCD4 were down-regulated in CCA tissues, compared with the matched normal tissues. HRP ×100
CCAs are aggressive cancers, with high mortality and poor survival rates. Only radical surgery offers patients some hope of cure; however, most patients are not surgical candidates because of late diagnosis secondary to relatively poor accuracy of diagnostic means.14,15Previous studies have demonstrated that microRNA-21 acts as an oncomir through complex regulatory network in multiple human cancers including CCAs.6,16-18However, the expression profile of microRNA-21 in CCA tissues and CCA cell line, QBC939 is not well established. In this study, we demonstrated microRNA-21 expression levels were increased by using real-time PCR and LNA-ISH analysis in CCAs tissues and cell line, compared with the normal tissues. These data suggest that elevated microRNA-21 expression is involved in cholangiocarcigenesis and microRNA-21 maybe plays the same role as in other cancers.
One of the best ways to understand miRNA function is via the elucidation of functional targets. This usually involves analysis of changes in target protein following either a gain- or loss-of function of the specific miRNA.19-22In this study, we not only demonstrate that PTEN and PDCD4 are bona fide targets of microRNA-21 in CCA cell line, QBC939 through blocking microRNA-21 function by using anti-microRNA-21, but indicate a negative correlation between microRNA-21 expression levels and its effectors, PTEN and PDCD4 proteins through LNA-ISH and immunohistochemistry analysis. These data provide strong evidence that PTEN and PDCD4 are direct targets of microRNA-21 in the formation and progression of CCA.
ACKNOWLEDGMENT
We thank Yi-hui Ma and Shuang-ni Yu for assistance with immunohistochemistry analysis.
1. Ambros V. The functions of animal microRNAs. Nature 2004; 431:350-5.
2. Hornstein E, Mansfield JH, Yekta S, et al. The microRNA miR-196 acts upstream of Hoxb8 and Shh in limb development. Nature 2005; 438:671-4.
3. Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature 2005; 436:214-20.
4. Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature 2005; 435:834-8.
5. Iorio MV, Visone R, Di Leva G, et al. MicroRNA signatures in human ovarian cancer. Cancer Res 2007; 67:8699- 707.
6. Selaru FM, Olaru AV, Kan T, et al. MicroRNA-21 is overexpressed in human cholangiocarcinoma and regulates programmed cell death 4 and tissue inhibitor of metalloproteinase 3. Hepatology 2009; 49:1595-601.
7. Papagiannakopoulos T, Shapiro A, Kosik KS. MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res 2008; 68:8164-72.
8. Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res 2005; 65:7065-70.
9. Lu Z, Liu M, Stribinskis V, et al. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene 2008; 27:4373-9.
10. Meng F, Henson R, Wehbe-Janek H, et al. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 2007; 133:647-58.
11. Gabriely G, Wurdinger T, Kesari S, et al. MicroRNA-21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol 2008; 28:5369-80.
12. Liu C, Yu J, Yu S, Lavker RM, et al. MicroRNA-21 acts as an oncomir through multiple targets in human hepatocellular carcinoma. J Hepatol 2010; 53:98-107.
13. Yamamichi N, Shimomura R, Inada K, et al. Locked nucleic acid in situ hybridization analysis of miR-21 expression during colorectal cancer development. Clin Cancer Res 2009; 15:4009-16.
14. Kerr TA, Davidson NO. Therapeutic RNA manipulation in liver disease. Hepatology 2009; 51:1055-61.
15. Mott JL. MicroRNAs involved in tumor suppressor and oncogene pathways: implications for hepatobiliary neoplasia. Hepatology 2009; 50:630-7.
16. Krichevsky AM, Gabriely G. miR-21: a small multi-faceted RNA. J Cell Mol Med 2009; 13:39-53.
17. Zhu Q, Wang Z, Hu Y, et al. miR-21 promotes migration and invasion by the miR-21-PDCD4-AP-1 feedback loop in human hepatocellular carcinoma. Oncol Rep 2012; 27: 1660-8.
18. Liu PT, Wheelwright M, Teles R, et al. MicroRNA-21 targets the vitamin D-dependent antimicrobial pathway in leprosy. Nat Med 2012; 18:267-73.
19. Guduric-Fuchs J, O'Connor A, Cullen A, et al. Deep sequencing reveals predominant expression of miR-21 amongst the small noncoding RNAs in retinal microvascular endothelial cells. J Cell Biochem 2012; 113:2098- 111.
20. Chau BN, Xin C, Hartner J, et al. MicroRNA-21 promotes fibrosis of the kidney by silencing metabolic pathways. Sci Transl Med 2012; 4:121ra18.
21. Zhang Q, Wang HY, Liu X, et al. IL-2R common gamma- chain is epigenetically silenced by nucleophosphin- anaplastic lymphoma kinase (NPM-ALK) and acts as a tumor suppressor by targeting NPM-ALK. Proc Natl Acad Sci U S A 2011; 108:11977-82.
22. Yao Q, Cao S, Li C, et al. Micro-RNA-21 regulates TGF- beta-induced myofibroblast differentiation by targeting PDCD4 in tumor-stroma interaction. Int J Cancer 2011; 128:1783-92.
