Association of Nurr1 gene mutations with Parkinson’s disease in the Han population living in the Hubei province of China**☆
Xiaoliang Lou, Weijing Liao
1 Department of Rehabilitation Medicine, Zhongnan Hospital, Wuhan University, Wuhan 430071, Hubei Province, China
2 Department of Neurology, Fourth Affiliated Hospital, Nanchang University, Nanchang 330003, Jiangxi Province, China
Association ofNurr1gene mutations with Parkinson’s disease in the Han population living in the Hubei province of China**☆
Xiaoliang Lou1,2, Weijing Liao1
1Department of Rehabilitation Medicine, Zhongnan Hospital, Wuhan University, Wuhan 430071, Hubei Province, China
2Department of Neurology, Fourth Affiliated Hospital, Nanchang University, Nanchang 330003, Jiangxi Province, China
Nurr1defects could in part underlie Parkinson’s disease pathogenesis, andNurr1gene polymorphism has been found in Caucasian patients with Parkinson’s disease. In this study, heteroduplex technology was applied to compare the DNA sequences of eight exons ofNurr1among 200 sporadic Parkinson’s disease patients and 200 healthy controls in the Han population in the Hubei province, China. One allele amplified from exon 3 ofNurr1was polymorphic in five Parkinson’s disease patients (2.5%, 5/200), and two individuals had a polymorphic allele amplified from exon 2 (1%, 2/200). The anomalous electrophoresis fragment in exon 3 ofNurr1gene contained a 709C/A missense mutation, and a polymorphic single nucleotide polymorphism at 388G/A was identified in exon 2. Compared with the control group, theNurr1gene expression level in the Parkinson’s disease group was decreased, and theNurr1gene expression levels in Parkinson’s disease patients carrying the polymorphisms at exons 2 and 3 were significantly decreased. Our data indicate that the single nucleotide polymorphism 388G/A in exon 2 and the 709C/A missense mutation in exon 3 of theNurr1gene in the Chinese population might affect the pathogenesis of Parkinson’s disease.
Nurr1gene; Parkinson’s disease; gene mutations; gene polymorphism; pathogenesis; neurodegenerative disease; neural regeneration
Research Highlights
(1) Heteroduplex analysis was used to conduct gene screening of eight exons ofNurr1gene among Parkinson’s disease patients and controls.
(2) Missense mutations in exons 2 and 3 of theNurr1gene were found in Chinese patients with Parkinson’s disease.
Abbreviations
DA, dopaminergic neurons; TH, tyrosine hydroxylase
lNTRODUCTlON
Parkinson’s disease manifests as many involuntary movement disorders, including tremor, reduced action and raised muscle tension. It is estimated that the number of Parkinson’s disease patients aged > 65 years in China is > 2 000 000, and the morbidity is increasing[1]. The molecular mechanisms underlying this disease are still not fully understood. A series of Parkinson’s disease-related genes have been identified, such as α-synuclein, Parkin 1-18 and others. However, the rare mutation rates of these genes still cannot account for most Parkinson’s disease molecular pathogenesis. Therefore, identification of gene polymorphisms controlling the production of dopaminergic neurons (DA), as well as their development and maintenance of function after maturity will provide new insights into the pathogenesis of Parkinson’s disease.
TheNurr1gene consists of eight exons and seven introns. The gene’s upstream 5’ flanking region contains a transcription promoter region and an adjusting zone. The first ATG codon from the 5’ end in exon 3 is the translation start point. A stop codon is located in the upstream region of exon 8, while the 3’ untranslated region of exon 8 contains multiple repetitive ATTTA sequences, and plays a stabilizing role in mRNA transcription.Nurr1is an immediate early gene and acts as gene transcription factor[2-4]. TheNurr1gene plays a dominant role in DA neuronal development, differentiation and the maintenance of function after maturation[5]. Zetterströmet al[6]reported that Nurr1 knockout rats died 1 day after birth, and that DA neurons were completely lacking in the ventral area of the brain. By contrast, tyrosine hydroxylase (TH) was detected in other regions, demonstrating that theNurr1gene plays a key role in maintaining the functions of mature DA neurons in the midbrain. Grimeset al[7]detected aNurr1gene polymorphism in the patients with Parkinson’s disease, and showed that theNurr1expression level was decreased by 45% in blood lymphocytes. Studies revealed that Parkinson’s disease patients carry exon 1 and intron 6 polymorphisms in theNurr1gene[8-9].
It was reported that theNurr1gene participates in the control of central dopamine metabolism[10], and that theNurr1expression level was significantly decreased with age in mesencephalic substantia nigra cells of the exon 3+/-heterozygous mouse, accompanied by decreased dopamine levels[11]. Nurr1 regulated TH metabolism and induced DA neuron formation. Transfection of embryonic stem cells with theNurr1gene can significantly improve their differentiation into DA neurons[12-14]. Thus, it can be speculated thatNurr1is a key gene for DA neuron development, differentiation and maintenance of functions, and that a lack ofNurr1will increase the environmental toxin sensitivity of neurons. Therefore, we used heteroduplex technology to conduct gene polymorphism analysis of eight exons of theNurr1gene in Parkinson’s disease patients from the Han population living in the Hubei province, China.
RESULTS
Quantitative analysis and clinical information of involved subjects
In this study, 200 sporadic Parkinson’s disease patients, including 110 males and 90 females (average age 62.03 ± 0.67 years) and 200 healthy controls consisting of 100 males and 100 females (average age 60.08 ± 0.82 years) were included. All subjects entered the final analysis. Clinical information for the Parkinson’s disease patients and healthy controls is shown in Table 1.

Table 1 Clinical characteristics of the participants
Exons 2 and 3 of the Nurr1 gene containing gene polymorphisms in Parkinson’s disease patients
We applied heteroduplex technology[15-19]to conduct fragment analysis of eight exons ofNurr1; abnormal electrophoresis fragments were subcloned and DNA sequencing was performed. We identified one allele amplified from exon 3 ofNurr1that was polymorphic in five Parkinson’s disease patients (5/200). Two individuals presented a polymorphic allele amplified from exon 2 (2/200), but no polymorphic allele existed in the healthy controls.
Gene mutation of exons 2 and 3 of Nurr1 in Parkinson’s disease patients
By DNA sequencing, we found that the anomalous electrophoretic fragment in exon 3 of theNurr1gene contained a 709C/A missense mutation (Figure 1). This mutation would change the 125thserine into tyrosine, affectingNurr1serine phosphorylation. However, further analysis needs to be carried out to determine the effect of the 388G/A polymorphism in exon 2 (Figure 2) onNurr1function, as exon 2 is part of theNurr1promoter region.
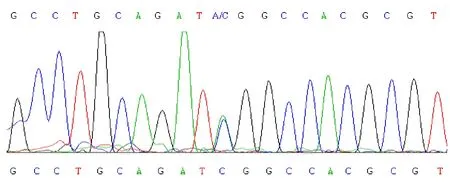
Figure 1 DNA sequencing diagram of 709C/A missense mutations in exon 3 of the Nurr1 gene.

Figure 2 DNA sequencing diagram of 388G/A missense mutations in exon 2 of the Nurr1 gene.
Nurr1 gene level decreased in Parkinson’s disease patients
Compared with the control group, theNurr1gene expression level of Parkinson’s disease group was decreased (P< 0.05). TheNurr1gene expression levels in Parkinson’s disease patients carrying the polymorphisms at exons 2 and 3 were significantly decreased compared with the control group (P<0.01; Table 2).
DlSCUSSlON
Nurr1is highly expressed in the developing and adult ventral midbrain and is required for the acquisition and maintenance of the dopaminergic phenotype in nigrostriatal neurons[20]. It has been reported that, besides the central nervous system,Nurr1is expressed in many other tissues including peripheral blood lymphocytes[21-23]. Using quantitative real-time PCR amplification[24-27], we analyzed theNurr1gene level in the peripheral blood lymphocytes of 200 sporadic Parkinson’s disease patients and 200 healthy controls and found that theNurr1expression level in thede novoParkinson’s disease group was significantly decreased compared with that in the healthy control group, especially in several patients carrying two polymorphisms in exons 2 and 3 ofNurr1, indicating that these mutations exert a down-regulating effect on the expression ofNurr1in peripheral blood lymphocytes. Studies on postmortem brains have found that an age-related decline in the levels of DA phenotypic markers is associated with down-regulation of Nurr1 expression in the human substantia nigra[28-29]. Chu and colleagues reported that the optical density of Nurr1 immunofluorescence was significantly decreased in nigral neurons containing α-synuclein-immunoreactive inclusions in Parkinson’s disease patients[30-31]. Therefore, we investigated whether theNurr1gene level in peripheral blood lymphocytes could exactly reflect the change in the disease stage in Parkinson’s disease patients. In the present study, we obtained similar results, similar to Le’s report that theNurr1gene level in human peripheral blood lymphocytes revealed a significant decrease in individuals with Parkinson’s disease and parkinsonian syndromes[32].
In conclusion, our present data showed that a single nucleotide polymorphism 388G/A in exon 2 and a 709C/A missense mutation in exon 3 of theNurr1gene exist in the Chinese population and might affect the pathogenesis in Parkinson’s disease patients.
SUBJECTS AND METHODS
Design
A gene polymorphic analysis.
Time and setting
The study was performed at the Zhongnan Hospital Affiliated to Wuhan University, China from January 2009 to October 2011.
Subjects
Parkinson’s disease patients were recruited from the Out-Patients Facility of Parkinson Clinic Center in Zhongnan Hospital, Wuhan University, China from 2009 to 2011.
Inclusion criteria
(1) Patients were diagnosed according to the UK Parkinson’s Disease Society Brain Bank Clinical Diagnostic Criteria for Parkinson’s Disease[33].
(2) All came from the Hubei Province of China.
(3) All patients were of Han ethnicity.
(4) Patients and their families signed an informed consent form.
Healthy physical examination controls were randomly selected from the Health Examination Center in Zhongnan Hospital, Wuhan University, China. In total, 200 sporadic Parkinson’s disease patients and 200 healthy controls were enrolled in this study, and matched for age, sex and ethnicity.
Methods
Nurr1 DNA analysis
Whole blood samples were collected from the upper limb veins of Parkinson’s disease patients and healthy controls. Genomic DNA was extracted from 4 mL of whole blood using the QIA-amp DNA Mini Kit (QIAGEN, Dusseldorf, Germany) according to the manufacturer’s protocols. Extracted genomic DNA samples were stored at -80°C until gene analysis was carried out. The primer pairs used to amplifyNurr1alleles were designed using the Primer3 (v. 0.4.0) system (PREMIER Biosoft International, Palo Alto, USA).
Nurr1PCR amplification primers and conditions are as follows:

Fragment Sequence (5’-3’) Annealing temperature (°C) Fragment size (bp) Exon1 Sense primers: CAT CTG TAC GCT CTT TCC GCT AA Antisense primers: CAT CCT TCG GTC CCA CTC T 59 430 Exon2 Sense primers: CAT ATG CCC AGC TGA ATC TC Antisense primers: GTT ACA GGG TTT GCC TTG TC 58 579 Exon3 Sense primers: TAA GGT TTG CCC GAC CCA TC Antisense primers: CTA CTG GCA CCA AGG CAG AG 60 305 Exon4 Sense primers: TTC TCC GAG TTG CCT GAT Antisense primers: TCC AAA TGG GTC GTA TAG TT 59 396 Exon5 Sense primers: TAA CAG GGC TCT TCC TTT GC Antisense primers: CCT TGC TTG CCT TCT TTA CC 59 487 Exon6 Sense primers: GCT GGA TGG CAC TGT ATT Antisense primers: AGC CTC CCT GGA TTG TCT 58 406 Exon7 Sense primers: ATG GAA TGG AGG TGG GAT AG Antisense primers: GTA CTG ACC TGT GAC CAT AG 60 438 Exon8 Sense primers: ATT GAT TCC ATT GTT GAA TTC TCC T Antisense primers: TGT GTA GTC CAT GTT CTA AAT CCA G 60 513
Briefly, eight exon-specific primer pairs were used to amplify exons 1-8 ofNurr1using a thermal cycler (model 9700, Applied Biosystems, Foster City, CA, USA). PCR products were electrophoresed through 4% acrylamide gels for 2 hours and abnormal alleles were sequenced using an ABI Prism 3100 Genetic Analyzer (Applied Biosystems). Sequence data were analyzed using Match Tools and Navigator software (Match Tools Allele Identification package, Applied Biosystems). The expression level ofNurr1gene was assessed as the average Ct value using real-time PCR in all Parkinson’s disease patients and healthy controls.
Statistical analysis
Data are expressed as mean ± SEM and were analyzed using SPSS 16.0 software (SPSS, Chicago, IL, USA). Comparisons between groups were performed using the chi-square test and Student’st-test.
Funding: This study was supported by the Science and Technology Department of Jiangxi Province, No. 20114BAB205076; and a Grant from the Jiangxi Provincial Health Department, No. 20094008.
Author contributions: Xiaoliang Lou was responsible for the data acquisition, and analysis, drafted the manuscript, conducted statistical processing, and was head of funds. Weijing Liao provided technical information, and supervised the experiment.
Conflicts of interest: None declared.
Ethical approval: The protocol for this study was approved by the Ethics Committee of Zhongnan Hospital, Wuhan University, China.
[1] Zhang ZX, Anderson DW, Huang JB, et al. Prevalence of Parkinson’s disease and related disorders in the elderly population of greater Beijing, China. Mov Disord. 2003; 18(7):764-772.
[2] Le W, Conneely OM, He Y, et al. Reduced Nurr1 expression increases the vulnerability of mesencephalic dopamine neurons to MPTP-induced injury. J Neurochem. 1999;73(5):2218-2221.
[3] Joseph B, Wallén-Mackenzie A, Benoit G, et al. p57(Kip2) cooperates with Nurr1 in developing dopamine cells. Proc Natl Acad Sci U S A. 2003;100(26):15619-15624.
[4] Zhang T, Wang P, Ren H, et al. NGFI-B nuclear orphan receptor Nurr1 interacts with p53 and suppresses its transcriptional activity. Mol Cancer Res. 2009;7(8):1408-1415.
[5] Saijo K, Winner B, Carson CT, et al. A Nurr1/CoREST pathway in microglia and astrocytes protects dopaminergic neurons from inflammation-induced death. Cell. 2009;137(1):47-59.
[6] Zetterström RH, Solomin L, Jansson L, et al. Dopamine neuron agenesis in Nurr1-deficient mice. Science. 1997; 276(5310):248-250.
[7] Grimes DA, Han F, Panisset M, et al. Translated mutation in the Nurr1 gene as a cause for Parkinson's disease. Mov Disord. 2006;21(7):906-909.
[8] Xu PY, Liang R, Jankovic J, et al. Association of homozygous 7048G7049 variant in the intron six of Nurr1 gene with Parkinson’s disease. Neurology. 2002;58(6):881-884.
[9] Le WD, Xu P, Jankovic J, et al. Mutations in NR4A2 associated with familial Parkinson disease. Nat Genet. 2003;33(1):85-89.
[10] Kim HJ. Stem cell potential in Parkinson’s disease and molecular factors for the generation of dopamine neurons. Biochim Biophys Acta. 2011;1812(1):1-11.
[11] Zhang L, Le W, Xie W, et al. Age-related changes in dopamine signaling in Nurr1 deficient mice as a model of Parkinson’s disease. Neurobiol Aging. 2012;33(5):1001.e7-16.
[12] Caiazzo M, Dell'Anno MT, Dvoretskova E, et al. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011;476(7359):224-227.
[13] Chang YL, Chen SJ, Kao CL, et al. Docosahexaenoic acid promotes dopaminergic differentiation in induced pluripotent stem cells and inhibits teratoma formation in rats with Parkinson-like pathology. Cell Transplant. 2012; 21(1):313-332.
[14] Johnson MM, Michelhaugh SK, Bouhamdan M, et al. The transcription factor NURR1 exerts concentrationdependent effects on target genes mediating distinct biological processes. Front Neurosci. 2011;5:135.
[15] Martini E, Borde V, Legendre M, et al. Genome-wide analysis of heteroduplex DNA in mismatch repair-deficient yeast cells reveals novel properties of meiotic recombination pathways. PLoS Genet. 2011;7(9):e1002305.
[16] Buitrago R, Depickère S, Bosseno MF, et al. Combination of cytochrome b heteroduplex-assay and sequencing for identification of triatomine blood meals. Infect Genet Evol. 2012;12(1):21-27.
[17] Mendonça MC, Ferreira AM, Santos MG, et al. Genotyping of human parvovirus B19 in clinical samples from Brazil and Paraguay using heteroduplex mobility assay, single-stranded conformation polymorphism and nucleotide sequencing. Mem Inst Oswaldo Cruz. 2011; 106(4):502-504.
[18] Jia X, Wei X, Liu Q, et al. An assay of gene copy number and its application based on heteroduplex products. Exp Mol Pathol. 2011;91(1):429-433.
[19] Cheng J, Yim OS, Low PS, et al. Detection of hemi/ homozygotes through heteroduplex formation in high-resolution melting analysis. Anal Biochem. 2011; 410(1):158-160.
[20] Kitagawa H, Ray WJ, Glantschnig H, et al. A regulatory circuit mediating convergence between Nurr1 transcriptional regulation and Wnt signaling. Mol Cell Biol. 2007;27(21):7486-7496.
[21] Liu H, Wei L, Tao Q, et al. Decreased NURR1 and PITX3 gene expression in Chinese patients with Parkinson's disease. Eur J Neurol. 2012;19(6):870-875.
[22] Le W, Pan T, Huang M, et al. Decreased NURR1 gene expression in patients with Parkinson's disease. J Neurol Sci. 2008;273(1-2):29-33.
[23] Satoh J, Nakanishi M, Koike F, et al. Microarray analysis identifies an aberrant expression of apoptosis and DNA damage-regulatory genes in multiple sclerosis. Neurobiol Dis. 2005;18(3):537-550.
[24] Heng X, Jin G, Zhang X, et al. Nurr1 regulates Top IIβ and functions in axon genesis of mesencephalic dopaminergic neurons. Mol Neurodegener. 2012;7:4.
[25] Eells JB, Wilcots J, Sisk S, et al. NR4A gene expression is dynamically regulated in the ventral tegmental area dopamine neurons and is related to expression of dopamine neurotransmission genes. J Mol Neurosci. 2012; 46(3):545-553.
[26] Nogueira EF, Rainey WE. Regulation of aldosterone synthase by activator transcription factor/cAMP response element-binding protein family members. Endocrinology. 2010;151(3):1060-1070.
[27] Spyroglou A, Manolopoulou J, Wagner S, et al. Short term regulation of aldosterone secretion after stimulation and suppression experiments in mice. J Mol Endocrinol. 2009; 42(5):407-413.
[28] Jacobs FM, van Erp S, van der Linden AJ, et al. Pitx3 potentiates Nurr1 in dopamine neuron terminal differentiation through release of SMRT-mediated repression. Development. 2009;136(4):531-540.
[29] McGeer PL, Itagaki S, Boyes BE, et al. Reactive microglia are positive for HLA-DR in the substantia nigra of Parkinson’s and Alzheimer’s disease brains. Neurology. 1988;38(8):1285-1291.
[30] Chu Y, Dodiya H, Aebischer P, et al. Alterations in lysosomal and proteasomal markers in Parkinson’s disease: relationship to alpha-synuclein inclusions. Neurobiol Dis. 2009;35(3):385-398.
[31] Chu Y, Mickiewicz AL, Kordower JH. α-synuclein aggregation reduces nigral myocyte enhancer factor-2D in idiopathic and experimental Parkinson’s disease. Neurobiol Dis. 2011;41(1):71-82.
[32] Le W, Pan T, Huang M, et al. Decreased NURR1 gene expression in patients with Parkinson’s disease. J Neurol Sci. 2008;273(1-2):29-33.
[33] Hughes AJ, Daniel SE, Kilford L, et al. Accuracy of clinical diagnosis of idiopathic Parkinson’s disease: a clinico-pathological study of 100 cases. J Neurol Neurosurg Psychiatry. 1992;55(3):181-184.
Cite this article as:Neural Regen Res. 2012;7(23):1791-1796.
Xiaoliang Lou☆, Studying for doctorate, Department of Rehabilitation Medicine, Zhongnan Hospital, Wuhan University, Wuhan 430071, Hubei Province, China; Department of Neurology, Fourth Affiliated Hospital, Nanchang University, Nanchang 330003, Jiangxi Province, China
Weijing Liao, M.D., Professor, Chief physician, Doctoral supervisor, Department of Rehabilitation Medicine, Zhongnan Hospital, Wuhan University, Wuhan 430071,
Hubei Province, China
Weijingliao@sina.com
2012-05-29
2012-07-27
(N20120327003/YJ)
Lou XL, Liao WJ. Association of Nurr1 gene mutations with Parkinson’s disease in the Han population living in the Hubei province of China. Neural Regen Res.
2012;7(23):1791-1796.
www.crter.cn
www.nrronline.org
10.3969/j.issn.1673-5374. 2012.23.005
(Edited by Liu SG, Gu T/Yang Y/Song LP)
- 中国神经再生研究(英文版)的其它文章
- Survey of spinal cord injury-induced neurogenic bladder studies using the Web of Science
- Current therapeutic strategies for inflammation following traumatic spinal cord injury☆●
- Stem cell therapy in neurodegenerative diseases From principles to practice●
- Meta-analysis of efficacy of topiramate in migraine prophylaxis★
- Differentiation of endogenous neural stem cells in adult versus neonatal rats after brachial plexus root avulsion injury*☆
- An enriched environment improves cognitive performance in mice from the senescenceaccelerated prone mouse 8 strain Role of upregulated neurotrophic factor expression in the hippocampus***☆

