Notch signaling dependent differentiation of cholangiocyte-like cells from rhesus monkey embryonic stem cells
JIN Li-Fang, JI Shao-Hui, YANG Ji-Feng,3, JI Wei-Zhi,*
(1. College of Life Science of Shaoxing University, Shaoxing Zhejiang 312000, China; 2. Kunming Institute of Zoology, the Chinese Academy of Sciences, Kunming Yunnan 650223, China; 3. College of Life Science of Wenzhou Medical College, Shaoxing Zhejiang 325035, China)
Notch signaling dependent differentiation of cholangiocyte-like cells from rhesus monkey embryonic stem cells
JIN Li-Fang1,2, JI Shao-Hui2, YANG Ji-Feng2,3, JI Wei-Zhi2,*
(1. College of Life Science of Shaoxing University, Shaoxing Zhejiang 312000, China; 2. Kunming Institute of Zoology, the Chinese Academy of Sciences, Kunming Yunnan 650223, China; 3. College of Life Science of Wenzhou Medical College, Shaoxing Zhejiang 325035, China)
Rhesus monkey embryonic stem (rES) cells have similar characteristics to human ES cells, and might be useful as a substitute model for preclinical research. Notch signaling is involved in the formation of bile ducts, which are composed of cholangiocytes. However, little is known about the role of Notch signaling in cholangiocytic commitment of ES cells. We analyzed the effect of Notch signaling on the induction of cholangiocyte-like cells from rES cells. About 80% of definitive endoderm (DE) cells were generated from rES cells after treatment with activin A. After treatment with BMP4 and FGF1 on matrigel coated wells in serum-free medium, rES-derived DE gave rise to cholangiocyte-like cells by expression of cholangiocytic specific proteins (CK7, CK18, CK19, CK20, and OV-6) and genes (GSTPi, IB4, and HNF1β). At the same time, expression of Notch 1 and Notch 2 mRNA were detected during cell differentiation, as well as their downstream target genes such as Hes 1 and Hes 5. Inhibition of the Notch signal pathway by L-685458 resulted in decreased expression of Notch and their downstream genes. In addition, the proportion of cholangiocyte-like cells declined from ~90% to ~20%. These results suggest that Notch signaling may play a critical role in cholangiocytic development from ES cells.
Rhesus monkey; Embryonic stem cells; Cholangiocytes; Notch signaling
Based on their ability to proliferate and their capacity to differentiate into specific cell types, embryonic stem (ES) cells are a potential source for cell transfer therapy in hepatic diseases. Although several studies have reported on the differentiation of hepatic cells from human (Duan et al, 2007), mouse (Hamazaki et al, 2001) and monkey ES cells (Ma et al, 2008; Jin et al, 2009), it remains unclear which factors are responsible for the induction of hepatic differentiation of ES cells.
Notch proteins are cell-surface receptors activated by interactions with cell surface ligands of the Jagged/Delta family. The mammalian family of Notch receptors consists of four members: Notch 1 through 4. Binding of the ligand to the Notch receptor induces site specific cleavage resulting in the release of the Notch Intracellular Domain (NICD). The NICD translocates to the nucleus where it modulates gene expression through interaction with members of the CSL (CBF-1, Suppressor of Hairless, lag-1) family of transcription factors. Activated Notch signaling elevates the expression of specific genes, including Hes 1 and Hes 5. Notch signaling has been shown to play an important role in cell-fate determination, cell survival, proliferation, and differentiation (Ma et al, 2007). The association of Notch signaling with cholangiocytes formation has been implicated in the literature (Lozier et al, 2008; Tanimizu & Miyajima, 2004); however, little is known about the mechanism of cholangiocytic commitment of ES cells.
In view of the close similarities between human and nonhuman primates, the rhesus monkey is a perfect animal model to better understand liver developmental processes in primates, and for preclinical assessment of cell transfer therapy protocols. In the present study, we analyzed the effect of Notch signaling on cholangiocytelike cells induction from rES cells in monolayer differentiation systems.
1 Materials and Methods
1.1 Culture of rhesus monkey ES cells
The rES cell line (366.4) was obtained from Dr. James Thomson, Wisconsin National Primate Research Center. Undifferentiated rES cells were expanded on a feeder layer of mitomycin-treated mouse embryonic fibroblasts seeded on 0.1% gelatin-coated plates, and cultured in Dulbecco’s modified Eagle medium (DMEM, Invitrogen, Fremont, CA) containing 15% FBS (Hyclone, Logan, UT), 1% nonessential amino acids (Sigma-Aldrich, Louis, MO), 0.1 mmol/L 2-mercaptomethanol (Sigma-Aldrich, St. Louis, MO), 2 mmol/L L-glutamine (Sigma-Aldrich, Louis, MO), and 1% penicillin/ streptomycin (Sigma-Aldrich, Louis, MO). Medium was changed daily, and rES colonies were manually split every four days to select for undifferentiated rES cells.
1.2 Growth factors induced cholangiocyte-like differentiation of rES cells
For definitive endoderm (DE) differentiation, the rES cell colonies were manually cut into small clumps of approximately 100 cells, plated on Matrigel-coated culture dishes, and differentiated in DMEM/F12 containing 1% FBS with 100 ng/mL human activin A (R&D systems) for 4-5 days. For cholangiocytic-like differentiation, the rES-derived DE cultures were passaged with 10 mg/mL of dispase and plated at a ratio of 1:2 on type I collagen (Sigma-Aldrich, Louis, MO) coated wells, and cultured in F12 medium supplemented with 2% knockout serum replacement (Gibco, Auckland, NZ), 100 ng/mL FGF1(R&D systems), and 20 ng/mL of BMP4(R&D systems) for 5−7 days. To study the effect of Notch signaling on cell differentiation, cells were cultured with or without 50 nmol/L Notch inhibitor (L-685458) (Bachem Bioscience, King of Prussia, PA) in F12 medium supplemented with 2% knockout serum replacement, 100 ng/mL of FGF1, and 20 ng/mL of BMP4 for 5−7 days.
1.3 Immunocytochemistry (ICC)
Cells were fixed with 4% paraformaldehyde in PBS for 10 min and washed with PBS three times, followed by permeabilization with 0.2% Triton X-100 for 10 min and blockage with 4% goat serum for 30 min at 25 °C. Subsequently, the cells were incubated with the primary antibodies (see Tab. 1) in staining solution (PBS containing 4% goat serum) for 40 min at 37 °C, and then incubated with the appropriate Texas red, PE or FITC-conjugated secondary antibody in staining solution for 30 min at 37 °C. The negative control was incubated in thestaining solution without primary antibodies.
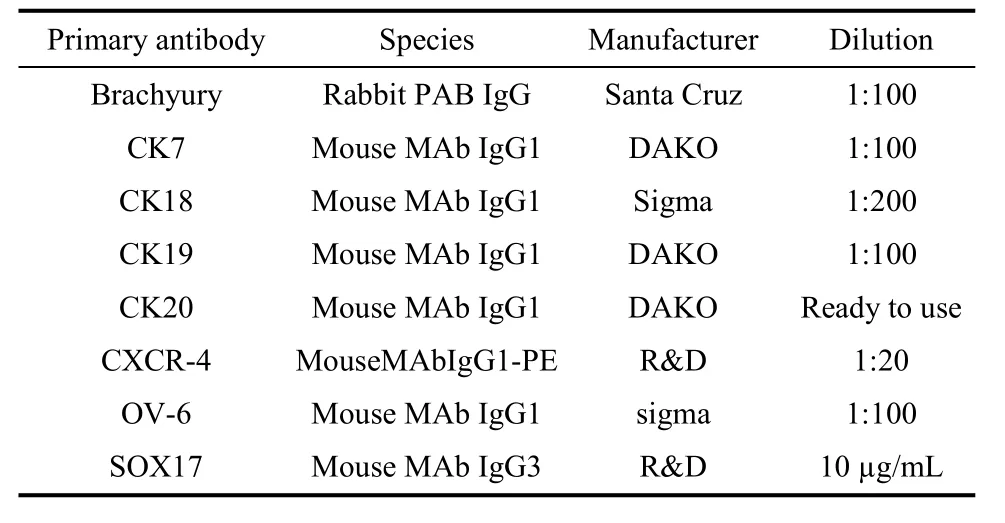
Tab. 1 Primary antibodies
1.4 Reverse transcription-polymerase chain reaction (RT-PCR)
Total RNA was isolated with Trizol reagent (Invitrogen, Calsbad, CA) according to the manufacturer’s protocols. Reverse transcription was carried out with approximately 1 μg of total RNA in 20 µL of 1 × master mix (1 × reverse transcription buffer, 0.5 mol/L dNTPs, 50 pmol of oligo(dT) primer, 20 U of RNase inhibitor and 5 U of reverse transcriptase) at 42 °C for one hour. For PCR, 1 µL of RT products was added to 1 × PCR master mix (1 × PCR reaction buffer, 1.5 mmol/L of MgCl2, 0.5 mmol/L of dNTPs, 0.4 µmol/L of forward primer, and 0.4 µmol/L of reverse primer (see supplemental Tab. 2), 1.25 U of Taq DNA polymerase in a 25 µL final volume and amplified by 25−35 cycles of PCR (95°C 30 s; 52 − 60 °C 30 s; 72 °C 30 s ) followed by a final extension at 72 °C for 5 min. All reagents in RT-PCR were purchased from Takara (Takara, Dalian, China) unless otherwise mentioned. Products of PCR were separated on a 2% agarose gel and stained with ethidium bromide. The primer sequence for IB4 is 5′-GCACGGACGAGATGTTCAG-3′/5′-ACTTGC CAAATCCAATAGTGTAG-3′; for GSTpi is 5′-GGACGG AGACCTCACCCTGTA-3′/5′-TCTTGCCTCCCTGGTTC TGG-3′; for HNF1β is 5′-GAAACAATGAGATCACTT CCTCC-3′/5′-CTTTGTGCAATTGCCATGACTCC-3′.
1.5 Analysis
For qualitative analysis, all differentiation experiments were replicated three times, andP≤0.05 was taken as significant.
2 Results and Discussion
2.1 Efficient generation of DE from rES cells
The rES clumps containing approximately 100 cells were collected and seeded on Matrigel-coated 4-well plates, and cultured with 100 ng/mL of activin A and 1% FBS for 5 days. After one day of induction, more than 90% of cells were strongly positive for brachyury, and only a few of the brachyury positive cells were faintly costained for SOX17(Fig. 1A). After two days of induction, the ratio of cells staining for both SOX17 and brachyury was beyond 80% (Fig. 1A). The SOX17/ brachyury double positive populations are regarded as generation of DE from ES cells (D'Amour et al, 2005; Yasunaga et al, 2005). In addition, the brachyury expression level gradually declined while the SOX17 expression level gradually increased with culture, with the peak of SOX17 expression observed at day four of differentiation (Fig. 1A), indicating that production of DE was a gradual differentiation process. On the contrary, untreated rES did not express brachyury and Sox17 (Fig. 1B).
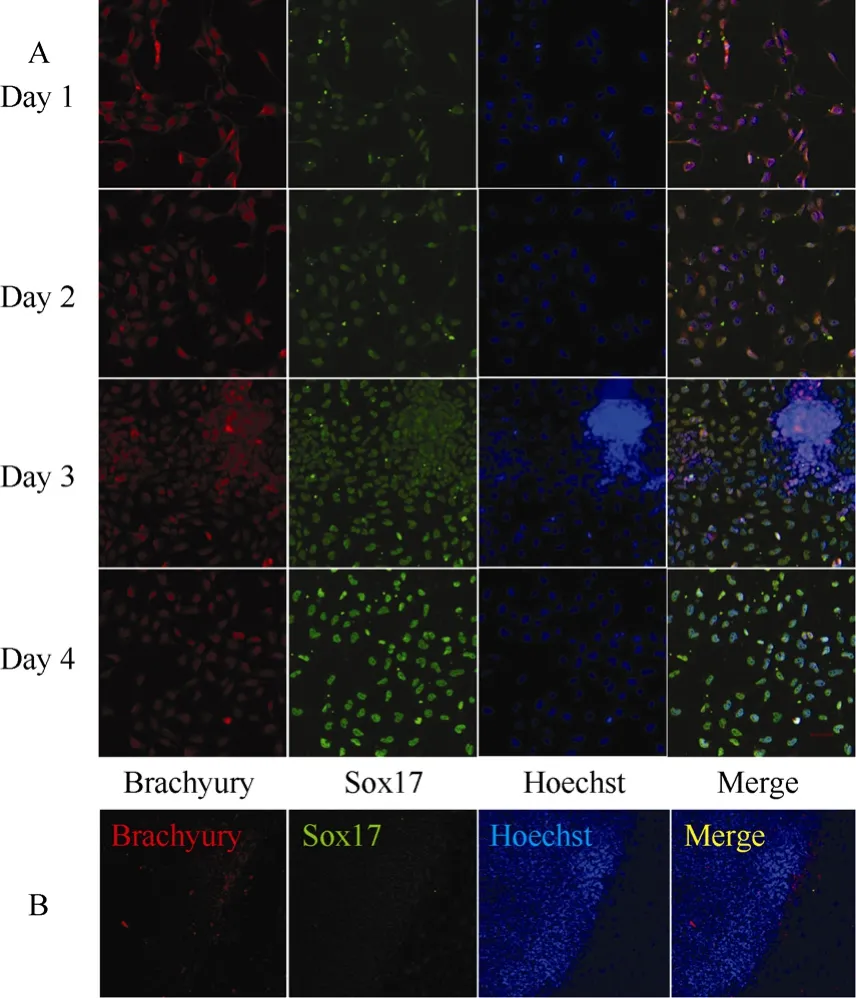
Fig. 1 Double labeling of differentiated cells by brachyury (red), SOX17 (green), Hoechst (nuclei, blue) and merged views of the same field of DE and rES cells
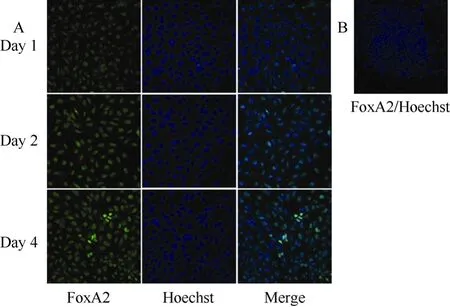
Fig. 2 FoxA2 was expressed in activin A induced rES cells
Consistent with the expression pattern of SOX17, we observed an increase in FoxA2 levels in activin A treated rES cells as differentiation proceeded (Fig. 2A), and untreated rES did not express FoxA2 (Fig. 2A). Genetic studies of nonmammalian organisms support a role for FoxA2 factors in endoderm development (Rehorn et al, 1996). Thus, expression of FoxA2 in differentiated cells also indicated that DE cells were generated.
To further verify that the SOX17 positive cells were DE cells, the CXCR4 expression was examined to distinguish DE from visceral endoderm (D'Amour et al, 2005). The CXCR4 positive cells were first observed on the third day of differentiation, and the proportion of CXCR4 positive cells increased up to 80% on the fourth day of differentiation (Fig. 3). Almost 100% of CXCR4 positive cells co-expressed SOX17 (Fig. 3). As previously mentioned, SOX17 was expressed in definitive, primitive and parietal endoderm cells, while CXCR4 was expressed in the mesoderm and DE cells. Thus, the SOX17/CXCR4 double positive populations should be definitive endoderm. Furthermore, the differentiated cells did not express the visceral endoderm marker alpha fetoprotein at any point during culture (data not shown). This evidence clearly indicates that activin A efficiently generated definitive endoderm from rES under low serum conditions.

Fig. 3 Double labeling of differentiated cells by CXCR4 (red), SOX17 (green), Hoechst (nuclei, blue) and merged views of the same field at 4 days
2.2 Cholangiocyte-like cells lineages differentiation of rES-derived definitive endoderm (DE) cells
Growth factors, such as FGFs, BMPs, HGF, OSM and Dex, are critical for hepatic endoderm morphogenesis and can promote hepatic differentiation (Kinoshita & Miyajima, 2002), and HGF, FGF-2 and OSM/Dex are critical for generation of hepatocytes from DEin vitro. In the present study, we examined the effect of BMP4 and FGF1 on hepatic fate differentiation from rES-derived DE. After 5−7 days induction, more than 90% of cultured cells expressed CK18 and CK19 (Fig. 4A), specific markers of bile duct cells. About 40% of cultured cells were stained for CK7 and CK20 (Fig. 4A), which are present in mature bile duct cells. In addition, another important protein of cholangiocytic marker OV6 was also detected in the differentiated cells (Fig. 4A). Conversely, DE cells were negative for CK7, CK20 and OV-6 (Fig. 4A). Cells were further analyzed by RT-PCR to confirm the presence of cholangiocyte-like cells in the differentiated cells. As expected, the differentiated cells expressed HNF1β, an important transcription factor controlling bile duct cell differentiation during liver development, as well as GSTPi and IB4, which are indicative of the presence of cholangiocyte-like cells (Fig. 4B). Unexpectedly, neither AFP nor ALB were detected in induced cells at either protein or mRNA levels. Thus, expression of the CK7, CK19, CK20 and OV6 proteins as well as the HNF1β, GSTPi and IB4 genes in cells from rES indicated that cholangiocyte-like cells were generated.
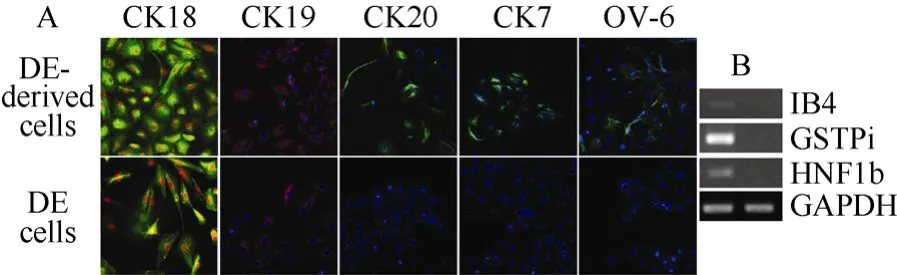
Fig. 4 Induction of cholangiocytes from rES-derived definitive endoderm by FGF1 and BMP4
2.3 Specification of rES-derived cholangiocyte like cells was Notch dependent
The Notch signaling pathway specifies cell fate during bipotential cell fate decisions and controls various biological events (Ehebauer et al, 2006), including the expression of transcription factors for exocrine cells, formation of bile ducts, and association with human Alagille syndrome (Crosnier et al, 2000). Notch 1 and Notch 2 are specifically expressed in proliferating hepatoblasts. Deletion of Notch 2 results in cholangiocyte and bile duct defects, while deletion of Notch 1 does not result in bile duct paucity (Lozier et al, 2008). Both the Hes1 and Hes5 genes, which encode a basic helix-loophelix protein, are downstream effectors of the Notch pathway. The Hes1-null mice formed a relatively-normal ductal plate consisting of cytokeratin- and DBA-positive cholangiocyte precursors, suggesting that the primary defect in these mice was not in the initial bipotential cell fate decision of the hepatoblasts (Kodama et al, 2004).
Given the importance of Notch activation in cholangiocyte-like specification from rES-derived definitive endoderm, we tested the expression of Notches and downstream target genes in the differentiation cultures. Notch 1 and Notch 2 mRNA were clearly detected after five days differentiation, as well as their downstream target genes such as Hes 1 and Hes 5 (Fig. 5B, line 1). The expression of Notch genes was accompanied by the expression of Notch intracellular domain (NICD) (Fig. 5A, untreated group). These results suggested that Notch signaling was active in cholangiocyte-like differentiation from rES-derived definitive endoderm.
To further confirm the role of Notch on cholangiocytelike specification, Notch inhibitor (L-685458) was supplemented in the differentiated medium at a concentration of 50 nM. After five days of culture, the expression of Notch genes as well as Hes 1 and Hes 5 significantly declined in the treatment group compared with the control group (Fig. 5B, line 2). As expected, the expression of NICD also declined (Fig 5B, treatment group). In addition, the proportion of CK19 positive cells significantly decreased to ~20% (as a percentage of total number of CK19-positive cells in multiple fields of hoechst positive cells), compared with ~90% of CK19 positive cells in the control group. Thus, consistent with previous studies (Kodama et al, 2004; Tanimizu & Miyajima, 2004), our results suggested Notch was essential for cholangiocyte-like differentiation from rES-derived DE.

Fig. 5 Notch signaling was critical for cholangiocytic differentiation from rES-derived definitive endoderm
In this study, we established reproducible and efficient protocols for definitive endoderm and cholangiocyte-like cell generation from rES cells by monitoring lineage development at different stages. Furthermore, our results suggested that the cholangiocytelike development of DE was Notch signaling dependent.
Crosnier C, Attie-Bitach T, Encha-Razavi F, Audollent S, Soudy F, Hadchouel M, Meunier-Rotival M, Vekemans M. 2000. JAGGED1 gene expression during human embryogenesis elucidates the wide phenotypic spectrum of Alagille syndrome [J].Hepatology, 32(3): 574-581.
D'Amour KA, Agulnick AD, Eliazer S, Kelly OG, Kroon E, Baetge EE. 2005. Efficient differentiation of human embryonic stem cells to definitive endoderm [J].Nat Biotechnol, 23(12): 1534-1541.
Duan Y, Catana A, Meng Y, Yamamoto N, He S, Gupta S, Gambhir SS, Zern MA. 2007. Differentiation and enrichment of hepatocyte-like cells from human embryonic stem cells invitroandin vivo[J].Stem Cells, 25(12): 3058-3068.
Ehebauer M, Hayward P, Arias AM. 2006. Notch, a universal arbiter of cell fate decisions [J].Science, 314(5804): 1414-1415.
Hamazaki T, Iiboshi Y, Oka M, Papst PJ, Meacham AM, Zon LI, Terada N. 2001. Hepatic maturation in differentiating embryonic stem cellsin vitro[J].FEBS lett, 497(1): 15-19.
Jin LF, Ji SH, Guo XY, Wang XH, Ji WZ. 2009. Induction of rhesus monkey embryonic stem cells into hepatocyte-like cells by a three-step method [J].Zool Res, 30(5): 509-514 (in Chinese).
Kinoshita T, Miyajima A. 2002. Cytokine regulation of liver development [J].Biochim Biophys Acta, 1592(3): 303-312.
Kodama Y, Hijikata M, Kageyama R, Shimotohno K, Chiba T. 2004. The role of notch signaling in the development of intrahepatic bile ducts [J].Gastroenterology, 127(6): 1775-1786.
Lozier J, McCright B, Gridley T. 2008. Notch signaling regulates bile duct morphogenesis in mice [J].PLOS One, 3(3): e1851, 1-6.
Ma A, Boulton M, Zhao B, Connon C, Cai J, Albon J. 2007. A role for notch signaling in human corneal epithelial cell differentiation and proliferation [J].IOVS, 48(8): 3576-3585.
Ma X, Duan Y, Jung CJ, Wu J, VandeVoort CA, Zern MA. 2008. The differentiation of hepatocyte-like cells from monkey embryonic stem cells [J].Cloning Stem Cells, 10(4): 485-493.
Rehorn KP, Thelen H, Michelson AM, Reuter R. 1996. A molecular aspect of hematopoiesis and endoderm development common to vertebrates andDrosophila[J].Development, 122(12): 4023-4031.
Tanimizu N, Miyajima A. 2004. Notch signaling controls hepatoblast differentiation by altering the expression of liver-enriched transcription factors [J].J Cell Sci, 117(15): 3165-3174.
Yasunaga M, Tada S, Torikai-Nishikawa S, Nakano Y, Okada M, Jakt LM, Nishikawa S, Chiba T, Era T, Nishikawa S. 2005. Induction and monitoring of definitive and visceral endoderm differentiation of mouse ES cells [J].Nat Biotechnol, 23(12): 1542-1550.
Notch 信号通路调控猕猴胚胎干细胞的胆管样细胞分化
金立方1,2, 纪少珲2, 杨纪峰2,3, 季维智2,*
(1.绍兴文理学院 生命科学学院,浙江 绍兴312000; 2.中国科学院昆明动物研究所 云南省动物生殖生物学重点实验室,云南 昆明650223; 3.温州医学院 生命科学学院,浙江 温州325035)
猕猴胚胎干细胞(rhesus monkey embryonic stem (rES))与人胚胎干细胞有相似的生物学特性, 因此是理想的临床前研究的替代模型。Notch信号通路在胆管及胆管上皮细胞的形成中有重要的作用, 然而, 有关Notch信号通路在ES细胞的胆向分化中的作用了解甚少。该实验以rES为模型, 对Notch信号通路对ES细胞的胆向分化过程中的作用进行了较为系统的研究。rES在细胞因子Activin A诱导作用下产生约80%的限定性内胚层细胞。以Matrigel作为细胞外基质, 在含BMP4和FGF1的无血清培养体系中继续诱导5~7 d, rES细胞来源的限定性内胚层细胞分化产生约胆管样细胞。分化的细胞表达胆管细胞的特异性蛋白((CK7、CK18、CK19、CK20 和OV-6)及基因(GSTPi、IB4 和 HNF1β)。在胆管样细胞的分化过程中检测到了Notch 1和Notch 2基因及下游信号分子hes 1和hes 5的表达。用Notch抑制剂L-685458处理分化过程中的细胞可导致Notch 1和Notch 2基因及下游信号分子hes 1和hes 5的表达下降, 同时CK19阳性的胆管样细胞分化比率也从90%下降至约20%。这一研究提示Notch信号通路可能在ES细胞的胆管样分化过程中扮演重要的角色。
猕猴; 胚胎干细胞; 胆管细胞; Notch 信号通路
Q813; Q959.848
A
0254-5853-(2011)04-0391-05
2011-01-21;接受日期:2011-06-11
金立方(1978-),男,浙江上虞人,讲师,博士,研究方向为干细胞生物学;E-mail: lifangj@sohu.com
10.3724/SP.J.1141.2011.04391
date: 2011-01-21; Accepted date: 2011-06-11
s: This work was supported by research grants from Zhejiang Natural Sciences Foundation of China (Y2110911; Y2080996), and the National Key Technologies R&D Program of China (2007CB947701)
*Corresponding author (通信作者), Tel/Fax: +86-0871-5139413, E-mail: wji@mail.kiz.ac.cn
- Zoological Research的其它文章
- 郑氏比蜢线粒体基因组全序列的测定与分析
- Localization of stationary pronuclei during conjugation of Paramecium as indicated by immunofluorescence staining
- 一种有效区分移植细胞和宿主细胞脑损伤模型的建立
- 悬尾应激对小鼠空间记忆及其反转学习的损伤效应
- Metabolism and thermoregulation between Mrs Hume’s Pheasant (Syrmaticus humiae) and Elliot’s Pheasant (S. ellioti)
- Afferent and efferent pathways in the visual system of the freshwater snail Planorbarius corneus

