Effects of Ionic Surfactants on Bacterial Luciferase and α-Amylase*
YAN Sangtian (闫桑田), LI An (李安), ZHENG Hao (郑浩), LUO Mingfang (罗明芳) and XING Xinhui (邢新会)**
Effects of Ionic Surfactants on Bacterial Luciferase and α-Amylase*
YAN Sangtian (闫桑田), LI An (李安), ZHENG Hao (郑浩), LUO Mingfang (罗明芳) and XING Xinhui (邢新会)**
Department of Chemical Engineering, Tsinghua University, Beijing 100084, China
In order to study the effects of ionic surfactants on bacterial luciferase, the cationic surfactant dodecyltrimethylammonium biomide (DTAB) and anionic surfactant sodium dodecylsulfate (SDS) were chosen. For comparison with bacterial luciferase, α-amylase was used since these two enzymes have similar electrostatic potential and charged active sites. After the enzymes were treated with the surfactants, the catalytic properties of bacterial luciferase and α-amylase were assayed, and fluorescence spectroscopy and circular dichroism (CD) were used to analyze the alteration of the protein structure. The results showed that when the DTAB concentration was low, the cationic surfactant DTAB enhanced the enzymatic activities of bacterial luciferase and α-amylase. On the other hand, the anionic surfactant SDS did not alter the enzymatic activity. The main interaction of cationic surfactant DTAB and the negatively charged surface of the proteins was the ionic interaction, which could alter the environment for the enzyme to work when the DTAB/enzyme molar ratio was low. However, at high cationic surfactant concentration, the ionic interaction and hydrophobic interaction might destroy the secondary and tertiary structures of the proteins, leading to the loss of enzymatic activities.
luciferase, α-amylase, ionic surfactant, surfactant-enzyme interaction
1 Introduction
Many organisms, ranging from bacteria and fungi to fireflies and fish, are capable of emitting light [1]. In these systems, bacterial bioluminescence (LUX) is generated by a complicated reaction catalyzed by bacterial luciferase, shown in Eqs. (1) and (2) [2]. The bacterial luciferase is encoded by theAB gene, which can be widely expressed in other organisms.
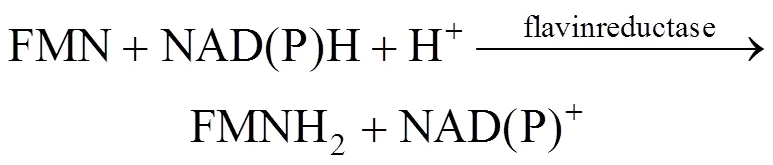

As seen from the above reaction mechanism, in addition to luciferase, intracellular coenzymes of FMN, FMNH2and NAD(P)H are also involved in luminescence. Bioluminescence is thus dependent upon bacterial cell concentration and metabolic activity. Bioluminescent bacteria have been used formonitoring of the toxicity of environmental samples and as commercial biosensors [3, 4]. Bioluminescence can also be used to indicate the biocatalyst concentration in mixed culture or in cofactor regeneration systems [5, 6]. In the above applications, the sensitivity of the bioluminescence formation is important. Recently, it has been reported that the light emission in the bioluminescence reaction by firefly luciferase is improved by a surfactant [7]. In our previous study [2], we use the cationic surfactant dodecyltrimethylammonium biomide (DTAB) to treat theBLU cells containing bacterialgenes, which significantly alters the bioluminescence output, but the reason remains unclear. Until now, few researches have been reported on analysis of the interaction between bacterial luciferase and surfactants.
Surfactant-enzyme (protein) interactions in aqueous solutions have been focused in many researches, and extensively studied for applications such as drug delivery, cosmetics and detergency, biodegradation, and for membrane proteins and lipids [8]. The interactions are important because they can modulate protein functions [9]. however, it is still very difficult to predict the molecular alteration of a protein by surfactants.
For interaction of surfactants with proteins, sodium dodecylsulfate (SDS) is the one studied most extensively. Other surfactants such as bile salts (sodium taurodeoxycholate, in particular), cetyltrimethylammonium bromide (CTAB) and DTAB [10] have also been investigated. The ionic head groups of the surfactants bind to the oppositely charged groups on protein surfaces by electrostatic interaction. The non-polar tail groups of the surfactants interact with the non-polar regions of proteins by hydrophobic interaction [9]. The intermolecular force of ionic surfactants and proteins involves electrostatic forces and polar molecule forces, and the electrostatic Coulomb force between the two charged atoms, or ions, is by far the strongest physical force [11].
In this study, in order to explore the effects of ionic surfactants on bacterial luciferase, the cationic surfactant DTAB and anionic surfactant SDS are chosen. The effects of surfactants on the enzyme structure and activity are examined. Since the surface amino acid charge in a protein is the key factor affecting the interaction between proteins and ionic surfactants [12], in addition to bacterial luciferase, several other enzymes including pepsin, firefly luciferase, α-amylase and lysozyme are used to estimate the surface electrostatic potential by Insight II software. The local charges on the protein surfaces are also very important, especially at the active site. According to the Insight II calculation results and the Protein Data Bank (PDB) database, the enzyme(s) with the most similar surface electrostatic potential and charges of the active site to those of the bacterial luciferase is chosen for comparison.
After the enzymes are treated with surfactants, the catalytic properties of bacterial luciferase and α-amylase are assayed. Since the interaction of ionic surfactants with enzymes may result in changes in the tertiary and secondary structures [15, 16], in this study fluorescence spectroscopy and circular dichroism (CD) are used to analyze the alteration of the protein structure.
2 Materials and methods
2.1 Materials
Two well-characterized enzyme proteins were used in this study: bacterial luciferase (EC 1.14.14.3; L8507; Sigma Chemical Co., molecular weight 40140) and α-amylase (EC 3.2.1.1; A4551; Sigma Chemical Co., molecular weight 44918). DTAB was purchased from TCI.-Decanol was purchased from Wako. SDS was purchased from Sigma. All other chemicals were of analytical grade and commercially available. The surface electrostatic potential of protein was calculated using the Insight II software (Biosym, San Diego, CA, USA, Accelrys Inc. 2001). The structural data of the two enzymes were downloaded from PDB [17], and input into the Insight II software to calculate the average surface electrostatic potential for a molecular structure under any pH condition. To choose the enzyme for comparison with the target bacterial luciferase, many enzymes including pepsin, bacterial luciferase, firefly luciferase, α-amylase and lysozyme were used for the molecular calculation.
2.2 Procedures
2.3 Analysis
2.3.1
The bacterial luciferase activity was evaluated by the bioluminescence (LUX) [1]. The reaction mixture for bioluminescence determination consisted of 1 ml.cell lysate, to provide the coenzyme for the reaction, and 50 µl 1 g·L-1bacterial luciferase. After the mixture was prepared and 10 µl-decanal was added, the bioluminescence was immediately detected by using a luminocounter (NU-700, Microteku-nichion, Japan). The waiting time and detection time of the luminocounter were set at 5 s, respectively [6]. The result of LUX was expressed by relative luminescence units (RLUs). Triplicate measurements were performed for each sample.
2.3.2
The fluorescence spectroscopy experiments were performed using a fluorescence spectrophotometer (F-2500, Hitachi Co., Japan). The enzyme solution after treatment with the surfactants was excited at 280 nm and the fluorescence due to the tryptophan and tyrosine residues was measured in the range of 300-500 nm. The value ofmaxfor the emission was found to be about 340 nm.
2.3.3
The changes in the secondary and tertiary structures of bacterial luciferase and α-amylase with the DTAB and SDS treatment under different conditions were evaluated by CD measurement. The far-ultraviolet CD spectra between 190 and 250 nm were recorded at 25°C with a spectropolarimeter (J-715, JASCO, Tokyo, Japan).
3 Results and Discussion
3.1 Enzymatic activity after treatment with DTAB and SDS
In order to compare the cationic surfactant DTAB and anionic surfactant SDS, the surfactant concentration, expressed by molar ratio (surfactant moles/enzyme moles), is in the range 100︰1 to 12000︰1 in this study. The enzymatic activity after treatment with different molar ratios of DTAB is shown in Fig. 1. At low DTAB concentration, the activities for the bacterial luciferase and α-amylase are enhanced. The highest LUX for the bacterial luciferase is 6.31×106RLU at molar ratio of 130︰1 (DTAB concentration 0.05 g·L-1), which is about 7-fold higher than the LUX of the control without the DTAB treatment. The α-amylase activity is also increased compared with the control, but the maximal increment is smaller than that of the bacterial luciferase. However, when the DTAB/enzyme molar ratio is higher than 1950︰1, the enzymatic activity of the bacterial luciferase decreases. The LUX is 3.87×105RLU at 1 g·L-1DTAB (DTAB/enzyme molar ratio 2600︰1), only about 44% of the control. Similarly, α-amylase keeps its improved activity until the DTAB/enzyme molar ratio reaches 2600︰1, and its activity decreases when the DTAB/enzyme molar ratio is higher than 2920︰1.
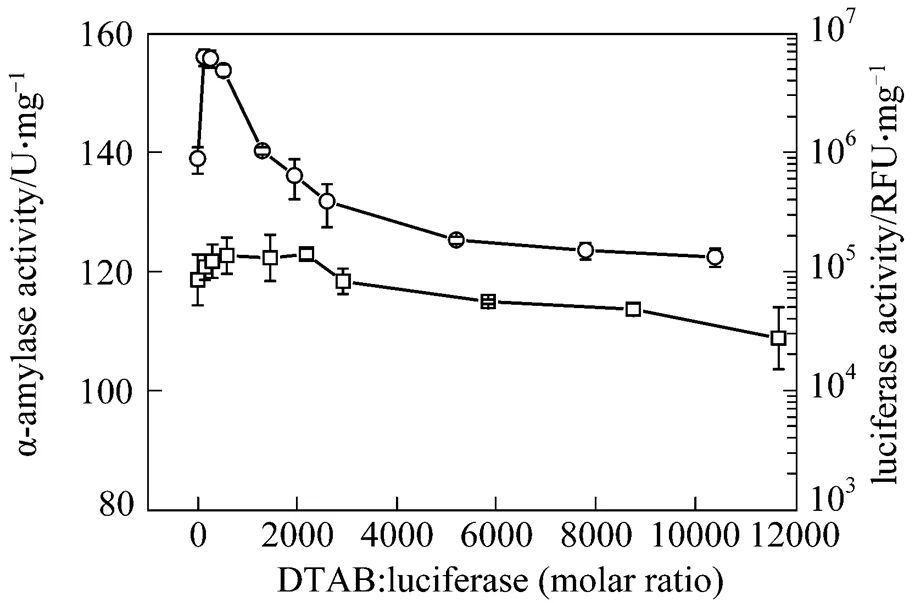
Figure 1 The activities of bacterial luciferase and α-amylase after treatment with DTAB for 20 min
□ α-amylase;○ bacterial luciferase
Figure 2 shows the enzymatic activity after treatment with SDS at different SDS/enzyme molar ratios. The anionic surfactant SDS does not enhance the activities of either bacterial luciferase or α-amylase. High SDS concentration also leads to a decrease of enzymatic activity for both enzymes. Moreover, in the control experiments, adding cofactor and surfactant without the luciferase does not affect the detection of LUX (data not shown).

Figure 2 Activities of bacterial luciferase and α-amylase after treatment with SDS for 20 min
□ α-amylase;○ bacterial luciferase
The above results indicate that the bacterial luciferase and α-amylase have the same behavior when treated with DTAB and SDS. For cationic DTAB,there is an optimal molar ratio of DTAB/enzyme for improving the enzyme activity, while the anionic SDS shows a different pattern.
3.2 Fluorescence spectroscopy
The fluorescence of bacterial luciferase and α-amylase depends upon tryptophan (Trp) residues (data not shown). Fluorescence spectroscopy is used to analyze the tertiary structure of the bacterial luciferase and α-amylase after treatment with DTAB and SDS (Figs. 3 and 4). As shown in Fig. 3, at low molar ratio of DTAB/enzyme (the ratio 200︰1 for bacterial luciferase and 234︰1 for α-amylase), the fluorescence intensities of these two enzymes are increased compared with the control. It is consistent withan investigation for another luciferase, firefly luciferase, in which the activity is enhanced by liposomes containing cationic surfactant at 0.5 pmol·L-1[7]. However, when the DTAB/enzyme molar ratio is higher than 260︰1 for the bacterial luciferase and 292︰1 for α-amylase, the enzymatic activities of both enzymes decrease.
Figure 4 shows that after treatment with SDS, the fluorescence intensity of the bacterial luciferase is increased when the SDS/enzyme molar ratio is less than 30︰1, and after that, as the ratio increases, the value ofmaxfor the emission of the bacterial luciferase becomes shorter (blue-shift) [Fig. 4 (a)]. However, the fluorescence intensity of α-amylase increases with the SDS/enzyme molar ratio in the whole range examined [Fig. 4 (b)].
The increase of the fluorescence intensity or the blue shift of the maximal emission wavelength implies that some parts of the enzymes are more hydrophobic, probably due to the exposure of the Trp to the environment. After treatment with a low molar ratio of DTAB/enzyme, the increases in the fluorescence intensity of the bacterial luciferase and α-amylase indicates that some parts of these two proteins become more hydrophobic [19, 20]. At higher DTAB/enzyme molar ratio, the obvious decrease in the fluorescence intensityindicates that the cationic surfactant changes the tertiarystructure of the bacterial luciferase and α-amylase, implyingthat more Trp residues and other amino acids areexposed to water, causing a more hydrophilic condition.
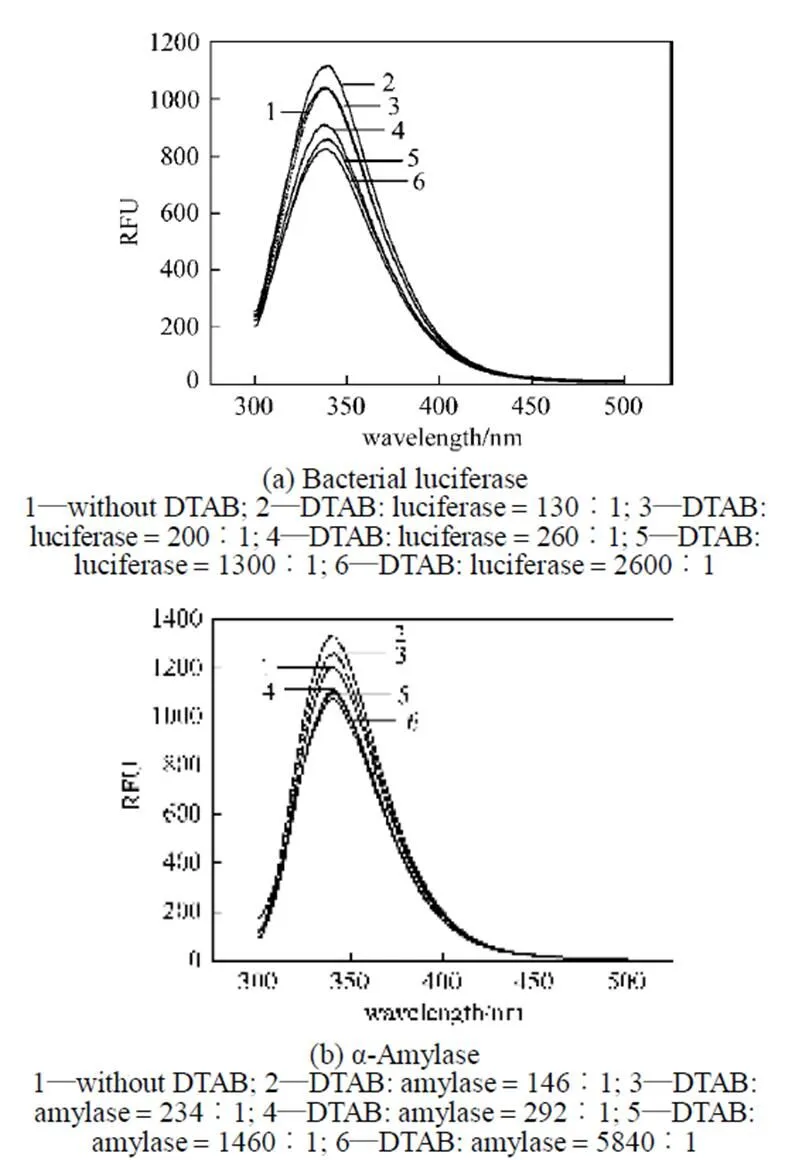
Figure 3 Fluorescence spectra of bacterial luciferase and α-amylase after treatment with DTAB for 20 min (EX wave 280 nm)
Figure 4 Fluorescence spectra of bacterial luciferase and α-amylase after treatment with SDS for 20 min (EX wave 280 nm)
In contrast, SDS, an anionic surfactant, can provide a more hydrophobic condition for these two enzymes [21]. The fluorescence spectroscopy of the bacterial luciferase and α-amylase after treatment with SDS suggests that SDS makes the enzyme surface more hydrophobic. For the bacterial luciferase, the fluorescence intensity increases with SDS concentration, but the maximal emission wavelength decreases. However, under the same conditions, for the α-amylase, the fluorescence intensity increases linearly with the SDS/enzyme molar ratio. The structure of α-amylase and bacterial luciferase is changed by SDS treatment, but α-amylase is more stable than bacterial luciferase in the presence of SDS.
3.3 CD spectra
The far-UV CD spectroscopy (190-250 nm) is shown in Figs. 5 and 6. When the DTAB/enzyme molar ratio is 130 for bacterial luciferase [Fig. 5 (a)] and 260 for α-amylase [Fig. 5 (b)], the CD spectra are not changed significantly, but when the molar ratio is increased to 1300 for the bacterial luciferase and 2600 for α-amylase, the CD spectra are changed obviously. In the CD spectra, the negative minimum peaks at around 209 nm and 222 nm reflect the α-helix [8, 19], and the negative minimum peak at 215 nm indicates the-sheet [19]. Fig. 5 indicates that after treatment with low DTAB/enzyme molar ratio, the α-helix of these two enzymes is destroyed first, and at higher DTAB concentration, the-sheet is destroyed.
After treatment with SDS at different concentrations, no significant changes occur in the CD spectra (Fig. 6), suggesting that the secondary structures of the enzymes do not change significantly after treatment with SDS in the range of SDS/enzyme molar ratios examined.
3.4 Interaction between the examined enzymes and surfactants
The effect of a surfactant on an enzyme’s structure and activity is dependent on the chemically selective interactions between the molecules, which may be influenced by enzyme structure and chemical property of surfactant. For many years, surfactants have been considered as non-specific denaturants of proteins, even though some positive effects of surfactants have been reported to enhance the activity and/or stability of some enzymes [12]. In this study, the interaction of the bacterial luciferase and α-amylase with cationic surfactant DTAB and anionic surfactant SDS are examined by evaluating the changes in the enzymatic activities and molecular structures.
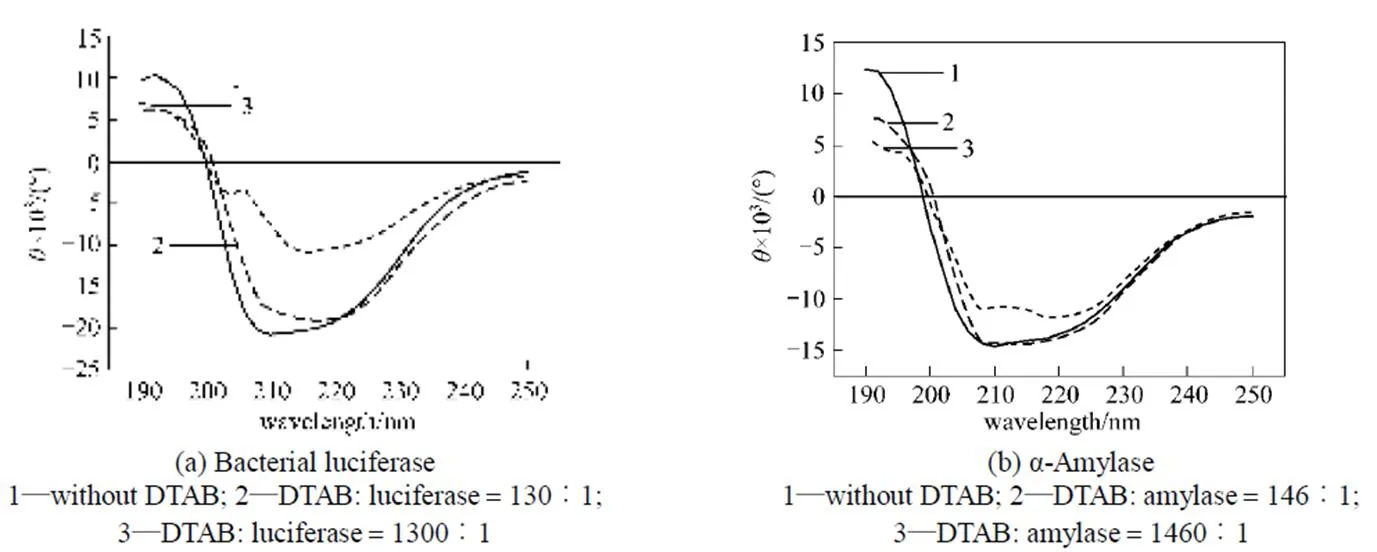
Figure 5 Circular dichroism spectra for the bacterial luciferase and α-amylase after treatment with DTAB
Figure 6 Circular dichroism spectra for bacterial luciferase and α-amylase after treatment with SDS
As shown in Figs. 1 and 2, when the DTAB concentration is low, the cationic surfactant DTAB may enhance the enzymatic activities of the bacterial luciferase and α-amylase, but the anionic surfactant SDS does not alter the enzymatic activities. It is widely accepted that the binding of ionic surfactant molecules to proteins can disrupt the native structure of most globular proteins [22-24]. Ionic surfactants interact with proteins through a combination of electrostatic and hydrophobic forces [25], so the surfactant head group will play a determining role in protein- surfactant interactions, which preferentially begins with the formation of strong ionic bonds between the surfactant polar groups, especially the charged sites on the protein surface [12]. Since the surface electrostatic potential and the local charges of the active sites on the protein surface of these two enzymes are similar at pH 7, the cationic surfactant DTAB may affect the surface anionic amino acid residues, which are negatively charged, or may bind to the negatively charged part on the surface in a similar pattern. The fluorescence spectrum analysis (Fig. 3) also indicates that when the DTAB concentration is low, some parts of the enzyme are exposed to a more hydrophobic environment. The electrostatic force between the active site and the surfactant may allow the active sites of the enzyme to become more flexible by changing its surrounding environment, increasing the enzymatic activity. At high DTAB concentration, the surfactant and protein association changes the enzyme structures, and the bacterial luciferase and α-amylase lose their activity, as indicated by the CD spectra (Fig. 5).
The main interaction of cationic surfactant DTAB and the negatively charged surface of the proteins is the ionic interaction, which can alter the environment in which the enzyme works when the DTAB/enzyme molar ratio is low. This interaction may contribute to the maximal luciferase activity at molar ratio of 130︰1 of DTAB to luciferase (Fig. 1). The detailed reason is still unclear now, but this phenomenon will be useful for further analysis of the DTAB-enzyme interaction mechanism. However, at higher cationic surfactant concentration, the ionic interaction and hydrophobic interaction may destroy the secondary and tertiary structures of the proteins, leading to the loss of enzyme activity.
On the other hand, the interactions of the bacterial luciferase and α-amylase with anionic SDS are different from those with DTAB. Since the surface electrostatic potential and especially the amino acid residues in the active sites of the two enzymes are negative, it may be difficult for the anionic surfactant to access the enzymes. Although the fluorescence spectra (Fig. 4) indicates that the enzymes are exposed to a more hydrophobic environment by in the presence of SDS, the protein structure does not change apparently in the range of the SDS/enzyme molar ratio examined, as reflected by the CD spectra (Fig. 6).
The results obtained in this study explain the phenomenon in which the luminescence of the bioluminescent.BLU is influenced by the addition of DTAB, and imply that DTAB may be used to enhance the luminescence of bacterial luciferaseor.
4 Conclusions
The effect of a surfactant on enzyme structure and activity is dependent on the chemically selective interactions between the two kinds of molecules, which may be influenced by the enzyme structure and the chemical property of the surfactant. When the DTAB concentration is low, the cationic surfactant DTAB enhances the enzymatic activities of bacterial luciferase and α-amylase, while the anionic surfactant SDS has little effect. At higher concentration of cationic surfactant DTAB, the surfactant and protein association may cause some changes in the enzyme structure. The ionic interaction and hydrophobic interaction may destroy the secondary and tertiary structures of the proteins, leading to the loss of the enzyme activity. The interactions of the bacterial luciferase and α-amylase with anionic SDS are different from those with DTAB. The protein structures are not changed apparently in the range of SDS/enzyme molar ratio examined.
1 Hastings, J.W., “Chemistries and colors of bioluminescent reactions: A review”,, 173, 5-11 (1996).
2 Tanaka, T., Xing, X.H., Matsumoto, K., Unno, H., “Preparation and characteristics of resting cells of bioluminescentBLU”,..., 12, 29-36 (2002).
3 Gil, G.C., Kim, Y.J., Gu, M.B., “Enhancement in the sensitivity of a gas biosensor by using an advanced immobilization of a recombinant bioluminescent bacterium”,.., 17, 427-432 (2002).
4 Lampinen, J., Virta, M., Karp, M., “Use of controlled luciferase expression to monitor chemicals affecting protein-synthesis”,..., 61, 2981-2989 (1995).
5 Burlage, R.S., Kuo., C.T., “Living biosensors for the management and manipulation of microbial consortia”,..., 48, 291-309 (1994).
6 Xing, X.H., Tanaka, T., Matsumoto, K., Unno, H., “Characteristics of a newly created bioluminescent pseudomonas putida harboring TOL plasmid for use in analysis of a bioaugmentation system”,.., 22, 671-676 (2000).
7 Kamidate, T., Niwa, S., Nakata, N., Application of cationic liposomes containing surfactants to an enhancer in firefly bioluminescent assay of adenosine 5′-triphosphate,.., 424, 169-175 (2000).
8 Hoshino,E., Tanaka, A., Kanda, T., “Effects of a nonionic surfactant on the behavior ofamyloliquefaciens alpha-amylase in the hydrolysis of malto-oligosaccharide”,.., 9, 63-68 (2006).
9 Kelley, D., Mcclements, D.J., “Interactions of bovine serum albumin with ionic surfactants in aqueous solutions”,, 17, 73-85 (2003).
10 Chakraborty, T., Chakraborty, I., Moulik, S.P., Ghosh, S., “Physicochemical studies on pepsin-CTAB interaction: Energetics and structural changes”,..., 111, 2736-2746 (2007).
11 Israelachvili, J.N., Intermolecular and Surface Forces, Academic Press, London (1991).
12 Savelli, G., Spreti, N., Di Profio, P., “Enzyme activity and stability control by amphiphilic self-organizing systems in aqueous solutions”,..., 5, 111-117 (2000).
13 Bordbar, A.K., Hosseinzadeh, R., Omidiyan, K., “Potentiometric study on interaction of dodecyltrimethylammonium bromide with alpha-amylase”,...., 77 (11), 2027-2032 (2004).
14 Bordbar, A.K., Hosseinzadeh, R., Omidiyan, K., “Study on interaction of alpha-amylase fromsubtilis with cetyl trimethylammonium bromide”,..-, 40, 2027-2032 (2005).
15 Gharibi, H., Javadian, S., Hashemianzadeh, M., “Investigation of interaction of cationic surfactant with HSA in the presence of alcohols using PFG-NMR and potentiometric technique”,..-..., 232, 77-86 (2004).
16 Wei, X.F., Liu, H.Z., “Relationship between foaming properties and solution properties of protein/nonionic surfactant mixtures”,.., 3, 491-495 (2000).
17 Berman, H.M., Battistuz, T., Bhat, T.N., Bluhm, W.F., Bourne, P.E., Burkhardt, K., Iype, L., Jain, S., Fagan, P., Marvin, J., Padilla, D., Ravichandran, V., Schneider, B., Thanki, N., Weissig, H., Westbrook, J.D., Zardecki, C., “The protein data bank”,..-.., 58, 899-907 (2002).
18 Stellmach, B., Bestimmungsmethoden Enzyme, Steinkopff Verlag, Darmstadt (1988).
19 Wu, D., Xu, G.Y., “Study on protein-surfactant interaction by spectroscopic methods”,..., 22, 254-260 (2006).
20 Zhao, N., Zhou, H., Biophysics, China Higher Education Press, Beijing (2000). (in Chinese)
21 Neu, T.R., “Significance of bacterial surface-active compounds in interaction of bacteria with interfaces”,.., 60, 151-166 (1996).
22 Bordbar, A.K., Moosavi-Movahedi, A.A., Amini, M.K., “A microcalorimetry and binding study on interaction of dodecyl trimethylammonium bromide with wigeon hemoglobin”,., 400, 95-100 (2003).
23 Goddard, E.D., Protein-Surfactant Interactions, CRC Press, New York (1993).
24 Sabate, R., Estelrich. J., “Interaction of alpha-amylase with-alkylammonium bromides”,...., 28 (2), 151-156 (2001).
25 Bordbar, A.K., Saboury, A.A., Housaindokht, M.R., Moosavi-Movahedi, A.A., “Statistical effects of the binding of ionic surfactant to protein”,..., 192, 415-419 (1997).
2009-03-03,
2009-06-03.
the National Natural Science Foundation of China (20676071, 20836004).
** To whom correspondence should be addressed. E-mail: xhxing@tsinghua.edu.cn
 Chinese Journal of Chemical Engineering2009年5期
Chinese Journal of Chemical Engineering2009年5期
- Chinese Journal of Chemical Engineering的其它文章
- Molecular Simulation of CO2/H2 Mixture Separation in Metal-organic Frameworks: Effect of Catenation and Electrostatic Interactions*
- Deactivation Kinetics of Nitrile Hydratase in Free Resting Cells*
- Corrosion Behavior of TP316L of Superheater in Biomass Boiler with Simulated Atmosphere and Deposit
- Influence of A-type Zeolite on Methane Hydrate Formation*
- Effects of Sintering Atmosphere on the Microstructure and Surface Properties of Symmetric TiO2Membranes*
- Improvement of Isomerization Process of Crude Isoamylene with Tertiary-amyl-alcohol Addition
