Lacticaseibacillus rhamnosus Probio-M9 may be vertically transmitted from mother to infant during lactation based on faeces metagenomics
Lan Yang,Lai-Yu Kwok,Zhihong Sun,Heping Zhang,
a Key Laboratory of Dairy Biotechnology and Engineering, Ministry of Education, Inner Mongolia Agricultural University, Hohhot 010018, China
b Key Laboratory of Dairy Products Processing, Ministry of Agriculture and Rural Affairs, Inner Mongolia Agricultural University, Hohhot 010018, China
c Inner Mongolia Key Laboratory of Dairy Biotechnology and Engineering, Inner Mongolia Agricultural University, Hohhot 010018, China
Keywords: Metagenomic analysis Lacticaseibacillus rhamnosus Gut microbiome Mother Infant
ABSTRACT Probiotics exert benef icial effects on the host.This study aimed to investigate whether maternally ingested Lacticaseibacillus rhamnosus Probio-M9 during pregnancy could access and colonize the infant gut.This study recruited one pregnant woman,who ingested Probio-M9 daily from 35 weeks of gestation to delivery.Feces of the mother-infant pair were regularly collected from one month before delivery to 6 months of infant’s age for metagenomic sequencing.Probio-M9 genomes were mappable to all infant fecal samples,suggesting the ingested probiotics could be vertically transmitted from mother to infant.Infant-or motherspecific differential metabolic pathways were found between the maternal and infant’s gut microbiome,implicating apparent differences in the intestinal metagenomic potential/function between the mother and the infant.In conclusion,maternal ingestion of Probio-M9 during the f inal weeks of gestation could deliver to the infant gut.The f indings provided novel insights into shaping infant’s gut microbiota.
1.Introduction
The human gut harbors trillions of symbiotic bacteria[1].Each individual has more than a hundred different gut microbial species.To date,about 1 500 different species have been identified as a part of human intestinal microbiota.There are 1013-1014microbes in intestinal microbiota[2-3],50 phyla and about 100-1 000 types of bacteria living in symbiosis,of which 150-170 types of bacteria are dominant,protecting the healthy human body[4].The stability of intestinal microecology and the metabolites directly affect the host health.The intestinal microecological preparations could accurately intervene in the intestinal microecology to improve host health.Microecological preparations are divided into probiotics,prebiotics and synbiotics,which have been widely used in health intervention[5].Probiotics are active microorganisms that exert benef icial effects on the host.Some common probiotics include the following categories:1)Bifidobacterium(Bifidobacterium longum,Bifidobacterium thermophilusand so on);2)Lactobacillus(Lactobacillus acidophilus,Lactobacillus,Lacticaseibacillus rhamnosusand so on);3) Gram-positive cocci (Streptococcus faecalis,Streptococcus thermophilusand so on);4) Fungi (yeasts and so on)[6].Some important physiological functions of probiotics include the regulation of the host intestinal microecological environment,suppression of pathogenic microorganisms in the gut,enhancement of epithelial barrier function,regulation of host immune response,and activation of internal immune cells with peptidoglycan metabolized by bacterial growth[7].
As the gut microbiota is closely linked with human health and the short-and long-term health impact of infant’s gut microbiota,active research has been undergoing in understanding the colonization and development of infant’s gut microbiota and its relationship with the maternal intestinal microbiota.Notably,after pregnancy,maternal microbiota undergoes changes and acquires specific signatures,and such changes might variably affect fetal development.The intestinal microbial community of the mother also directly affects the establishment of the infant’s gut microbiota[8].One previous study found that supplementingL.rhamnosusGG (LGG,1.8 × 1010CFU/day) from 35 weeks of gestation to delivery could promote the colonization ofBifidobacteriumin breastfed infants’gut[9].Moreover,the mother was able to transfer LGG to the baby,promoting its gut bifidobacterial diversity[10].Thus,ingesting probiotics during pregnancy can regulate the intestinal microbiota of both the mother and the infant.
High-throughput sequencing is a powerful method enabling metagenomics analysis that covers both the cultivable and uncultivable microbial populations in samples.Metagenomic sequencing has been used to describe the taxonomic composition and functional potential of microbial communities,as well as to assemble the entire genome sequence of a specific strain[11].Metagenomic data analysis has also enabled us to gain a comprehensive understanding of microbial diversity,composition,community function,and their relationship with the colonic environment.Cost-effective and highthroughput metagenomic techniques have been widely used,and deep phylogenetic characteristics of the intestinal microbiome have been successfully analyzed.Metagenomic sequencing technology can be used to assess the impact of probiotic consumption on gut microbiota composition,structure,and function[12].The composition of intestinal microbiota varies greatly between individuals with relatively conserved functions[13].However,some factors,such as probiotic administration,might influence the gut microbiota diversity and composition,maintaining a healthy stability of the physiological state of the gut[14-15].Therefore,it is of interest to investigate the beneficial effects of probiotic consumption on intestinal microbiota composition and function.
L.rhamnosusProbio-M9 (Probio-M9) is a probiotic independently isolated from the breast milk of a healthy mother in China.Probio-M9 has been shown to maintain the homeostasis of host intestinal microbiota and was good[16].This work aimed to investigate 1) whether Probio-M9 could transmit vertically from mother to infant,and 2) the effects of Probio-M9 consumption on the microecological and microbiota functions of maternal and infant intestines.In this study,one pregnant woman ingested Probio-M9 from 35 weeks of gestation until delivery,then the faecal microbiota structure and function of the mother-infant pair was studied by metagenomic methods.Our results supported that Probio-M9 intake during gestation could be vertically transmitted from mother to infant,and it could modulate the gut microbiota structure and function of intestinal microbe of both the mother and the infant.
2.Materials and methods
2.1 Ethics statement
This work was authorized by the Ethics Committee of Inner Mongolia Medical University (under the registration number ChiCTR2100044607,Chinese Clinical Trial Registry).The participant provided written informed consent prior to starting the study.
2.2 Subject recruitment
Pregnant woman was recruited by the Inner Mongolia Agricultural University.One normal primigravida at 35 weeks of gestation,normal BMI,no periodontal disease,no type 2 diabetes,no vaginitis,and without other histories of serious/chronic diseases was selected.The mother did not take antibiotics during pregnancy and lactation,and the fetus was delivered by caesarean section at full term.The participant underwent an ultrasound examination at term to assess the state of fetal wellbeing.
2.3 Study design and sample collection
The pregnant woman was given probiotic supplements soon after providing her written consent for trial participation and receiving instructions for probiotic intake and sampling.The participant was requested not to take antibiotics and other probiotic-containing foods during the trial period and to notify her doctor of any abnormalities.The participant was also asked to return the probiotic packaging materials for compliance assessment.
The pregnant woman received 2 g (2.5 × 1010CFU/g) of Probio-M9 powder after dinner every evening,and the intervention started 30 days before the expected date of confinement until the day of delivery.The probiotics were provided by the Key Laboratory of Dairy Biotechnology and Engineering,Ministry of Education,Inner Mongolia Agricultural University.The first sample was collected 2 days before starting the first probiotic administration (D-30) and was performed by trained professionals under strict aseptic conditions using standard procedures.Meanwhile,the participant was taught the standard sampling method.About 10 g of faeces sample was collected each time.From the first sample collection to the day of delivery,faeces samples were collected every 7 days counting from the first probiotic intake (D-28).Sampling was continued after the delivery from the mother and the infant.For the first week after delivery,samples were collected on the day of delivery (D0) and days 1,2,3,and 7 after delivery (D1,D2,D3,and D7,respectively).From the 2ndweek to 2thmonths after delivery (D14 to D56),faeces samples were collected every 7 days.From the 3rdto 6thmonth after delivery(D70 to D182),sampling was done every 14 days.The complete sampling lasted 212 days;however,as the participants did not defecate at certain time points,only a total of 33 faecal samples were collected from the mother (20 samples) and the infant (13 samples),respectively (Fig.1).

Fig.1 The timing of collecting faeces samples from mother and infant.
Faeces samples were collected into sterile tubes provided by our laboratory beforehand.Collected samples were stored temporarily in a household refrigerator (-20 °C) and were transported to the laboratory by our staff within 24 h of sample collection.They were stored at-80 °C until total DNA extraction.
2.4 DNA extraction
Metagenomic DNA was extracted from faeces samples using QIAamp Fast DNA Stool Mini-kit (QIAGEN,Hilden,Germany).The quality of extracted DNA quality was evaluated by agarose gel electrophoresis and NanoDrop spectrophotometry (optical density ratio at 260 nm/280 nm).All DNA samples were stored at -20 °C until further processing.
2.5 Whole genome metagenomics sequencing and quality control
Metagenomic libraries containing 2 µg of genomic DNA were constructed according to the Illumina TruSeq DNA Sample Prep V2 Guide.The qualities of all libraries were assessed by an Agilent bioanalyzer and the DNA LabChip 1000 Kit.Sequencing was performed on an Illumina HiSeq XTEN sequencer (Illumina,San Diego,USA).The quality control filters were set according to Liu et al.[17].1) Reads with adaptor sequences were removed by the software SeqPrep.2) Reads were trimmed from the 3’ end using a quality threshold of 30.3) Low quality (Q30) reads comprised more than 50%of the bases were removed.4) Reads less than 70 bp were removed by Sickle software.5) Reads of host genome sequences were removed.High-quality reads (a total of 363.45 Gb,average of 11.01 Gb per sample) were retained and used for the further analysis.
2.6 Bioinformatic analysis
MetaPhlAn3 (Ver.3.0) was used for species-level taxonomic assignment of faeces microbiota using default settings via the search engine Bowtie2 (Ver.2.2.9)[18-19].PanPhlAn3 (Ver.3.0.1)was used for species-level analysis[20].If a strain was identified by PanPhlAn3,the abundance of the strain would be regarded as the species abundance.High-quality Illumina sequencing-generated metagenomic datasets were assembled by MegaHit (Ver.1.0)[21].QUAST (Ver.5.0.0) was used to evaluate the metagenomic assembly results.MetaBAT2 (Ver.2.12.1) and Maxbin2 (Ver.2.0)were used for binning the assemblies using a minimum scaffold length threshold of 1 500 bp[22-23].Das Tool (Ver.1.1.2) was used to connect the contigs assembled by MetaBAT2 and Maxbin2[24].Metagenome-assembly genomes (MAGs) in conformity with quality requirements (integrity > 80%,contamination < 10%) were selected.CheckM (Ver.1.0.18) was then used to evaluate the integrity and contamination degree of each MAG[25],and high-quality MAGs were used for further analysis.
Each MAG was annotated by BLASTn using the Non-redundant Nucleotide Sequence Database (NT) of National Center for Biotechnology Information (NCBI),phylogenetic trees were constructed using 400 generic PhyloPhlAn markers,and the MAGs were clustered by dRep (Ver.2.2.4) to define species-level genome bins (SGBs)[26].
2.7 Statistical analysis
R package vegan was used for alpha and beta diversity analyses.Statistical differences in alpha diversity were assessed by Wilcoxon rank-sum tests.Multivariate analysis,such as principal coordinate analysis (PCoA),was performed by R software (Ver.4.0.3;https://www.rproject.org/).Differential metabolic pathways between mother’s and infant’s gut microbiome were identified byt-tests.Correlations between different biological indicators were assessed with the Spearman’s rank correlation coefficient and visualized by Cytoscape 3.5.1.Spearman’s correlation analysis was also performed to find significant correlations between the microbial community and metabolites (r> 0.6,P< 0.05).
To identify Probio-M9 in samples,metagenomic DNA from all samples were mapped to the reference genome of Probio-M9(comment: add genome database,accession number).To avoid ambiguity alignment,only reads with alignment similarity of > 95%were included in the analysis.The strain was considered to be present in a specific metagenomic sample if the coverage of the reference(Probio-M9) genome was greater than 40%[27].
2.8 Nucleotide sequence accession numbers
Datasets generated by the whole genome metagenomics sequencing in this study were deposited in the NCBI Short Read Archive (SRA) database under accession number PRJNA770101,http://www.ncbi.nlm.nih.gov/bioproject/770101.
3.Results
3.1 Matching between faecal metagenomic samples of mother-infant pair with reference genome
High-quality reads of our dataset (20 and 13 samples from the mother and the infant,respectively) were assembled and mapped to the reference genome of Probio-M9 by Blastn.The matching portions between metagenome of each maternal and infant sample with the reference genome are shown in Fig.2A.Notably,the genome coverage breadth of all 13 infant metagenome matched > 40% with the Probio-M9 reference genome,contrasting to only one positive match (i.e.,AM10 from D42 after delivery;Fig.2B).
3.2 Microbial dynamics of mother’s and infant’s faecal microbiomes
The dynamics of the faecal microbiome composition and diversity of the mother-infant pair was tracked over a period of 212 days.Species-level taxonomic annotation was performed with MetaPhlAn3.The faecal microbiota of all mother and infant together contained sequences representing 303 species that belonged to six phyla and 51 families.There were 15 major species in the complete dataset (average relative abundance > 1.0%;Fig.3A).Generally,the microbial complexity was rather low in the infant’s faeces with only two major species,includingBifidobacterium breveandBifidobacterium pseudocatenulatum.In contrast,the mother’s faecal microbiota was relatively complex,comprising a number of major species,e.g.,Bacteroides stercoris,Bacteroides vulgatus,Eubacterium rectale,Faecalibacterium prausnitzii,B.pseudocatenulatum,andBacteroides uniformis.
The metagenomic data were subjected to PCoA (Bray-Curtis distance;Fig.3B).Symbols representing the mother’s and infant’s microbiota showed distinct clustering patterns on the PCoA score plot,confirming great differences in the faecal microbiota between the two individuals and little variation among samples of the same person.Neither mother’s nor infant’s samples exhibited time-based trends on the PCoA score,suggesting that probiotic treatment did not cause any obvious temporal changes.While the correlation between the gut microbiota of mother and infant was worth exploring.
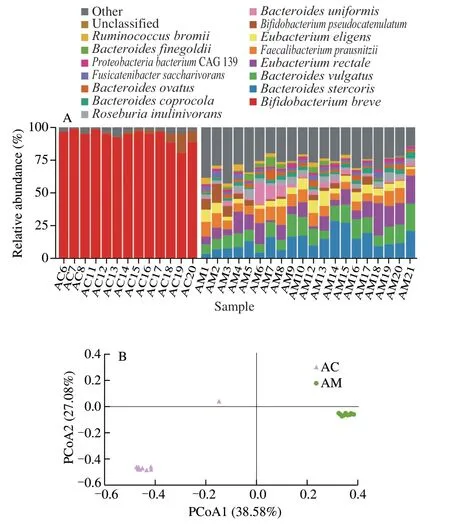
Fig.3 Faecal microbiota composition and diversity of the mother-infant pair.(A) Faecal microbiota composition.Non-major species of relative abundance <1% of the total sequences are grouped under ‘others’.(B) PCoA (Bray-Curtis distance) of mother and infant faecal microbiota.The codes,AC and AM,represent samples of the infant and the mother,respectively.
3.3 Effect of Probio-M9 intake on correlation network of faecal microbiota of mother and infant
A correlation network of faecal microbiota of the mother and infant was constructed to explore the impact of Probio-M9 on their gut microbiota (Fig.4).The top 30 species were included in the correlation analysis.Interestingly,most species showed significant positive correlation (r=0.8,P< 0.05),while a few species exhibited significant negative correlation (r=0.8,P< 0.05).B.breve(predominantly present in infant’s sample)showed significant negative correlation with some bacteria of mother,such asB.vulgatus,E.rectale,F.prausnitzii,Eubacterium eligens,B.uniformis,Roseburia inulinivorans,andFusicatenibacter saccharivorans.These results showed that there was a complex correlation between mother’s and infant’s gut microbiota.
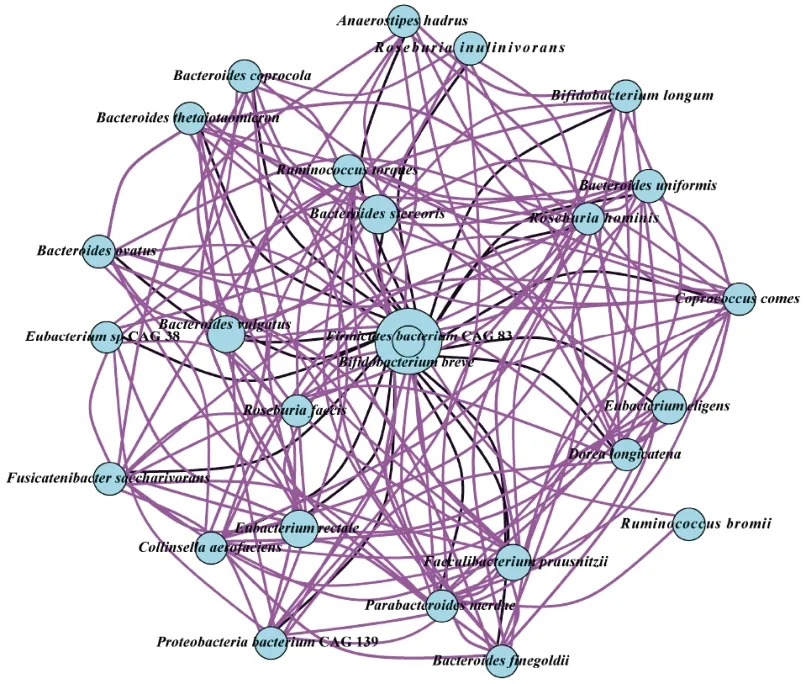
Fig.4 Correlation network between faecal microbiota of the mother and the infant.The top 30 most abundant features were used to construct the network.The size of the circle represents the relative abundance of the specific species.Positive and negative Spearman’s correlations were represented by purple and black lines,respectively.The line thickness represents the strength of correlation as illustrated by the color scheme.The color scheme represents the Spearman’s rho,ranking between 0.8 and -0.8.P value was less than 0.05.A value greater than 0 indicates positive correlation,and vice versa.
3.4 Metagenomic potential of mother’s and infant’s microbiota
The metagenomic potential of mother’s and infant’s gut microbiota was annotated,and differential metabolic pathways were identified between the mother’s and infant’s data subsets (Fig.5).It is interesting to note that the gene abundance of these metabolic pathways was relatively stable in the mother’s but not the infant’s microbiome over time.The results of cluster heatmap broadly classified these metabolic pathways into three groups.The first group comprised 9 metabolic pathways,including PWY-7204,PWY0-1241,and PWY-7269.Genes coding these metabolic pathways were mainly detected in the infant’s samples.The second group contained 8 metabolic pathways,including P562-PWY,PWY-6863,and P162-PWY.Genes coding these metabolic pathways were mainly observed in the maternal samples,but with a relatively low amount.The third category possessed 11 metabolic pathways,including PWY-5676,GALACT-GLUCUROCAT-PWY,and PRPP-PWY.These pathways were also mainly encoded in the maternal faecal microbiome.However,their relative contents were relatively high compared with those in the second group.These results indicated obvious differences existed in the metagenomic potential between the maternal and infant gut microbiota,suggesting different physiological conditions within the maternal and infant’s gut,which might in turn shape the functions of their microbial communities,respectively.
4.Discussion
The gut microbiota is closely related to human health and nutrition.Gut dysbiosis may predispose the host to a wide spectrum of chronic medical conditions,including metabolic diseases,dietary and nutritional problems.Probiotics have been shown to effectively improve human health and gut homeostasis.A number of clinical trials and metagenomic sequencing analyses have been performed to explore the function of probiotics in improving the human gut microbiota.This study used metagenomic sequencing to investigate how maternal intake ofProbio-M9 influenced the composition of maternal and infant’s gut microbiota,the interaction between intestinal microbiota,and the function of intestinal microbiota.
L.rhamnosusis one common probiotic species that has been safely used in various functional foods for nearly 30 years[28].It has many health effects,such as preventing and relieving certain types of diarrheas[29],regulating inflammatory response[30],and lipid metabolism[31].In previous studies,LGG has been well known for its health benefits,but otherL.rhamnosusstrains with health benefits are in urgent need of development.Probio-M9 is aL.rhamnosusstrain isolated from breast milk of a healthy woman,which has been shown to possess health-promoting functions to human.The purpose of this study was to explore whether Probio-M9 could be transmitted from pregnant mother to infant,and its effects on gut microbiota of mother and infant.

Fig.5 Cluster heatmap of differential metabolic pathways between the maternal and infant’s faecal microbiota.The 28 significant differential metabolic pathways were identified by t-test (cut-off: P < 0.05) and log2|fold change| > 1 between metabolites of the mother-infant pair.The codes,AC and AM,represent samples of the infant and the mother,respectively.
L.rhamnosuscolonizes the gut and is fundamental to human health.Our results showed that maternal oral intake of Probio-M9 could deliver the probiotic bacteria to the infant’s gut.Interestingly,Probio-M9 was successfully detected throughout the trial period until the infant reached the age of 6 months,even though the maternal intake of Probio-M9 ceased at the day of delivery.These results suggested that the maternal dietary intake of probiotics not only could be vertically transmitted from mother to infant,but these bacteria were likely able to colonize the infant’s intestine during the trial period.On the other hand,Probio-M9 was not detected in the maternal faecal samples,which could be due to the detection limit (sampling volume,insufficient sequencing depth and so on) of the metagenomic sequencing method for specifically detecting the maternally ingested Probio-M9,which was of relatively low quantities relative to the large number of bacteria present in adults’ colon.Moreover,the infant gut microbiota is generally considered to be of low diversity and often comprises a relatively high amount of bifidobacteria,making it more feasible to detect a specific target strain in infant than adult infant faecal samples[32].
The development of intestinal microbiota during the first 1 000 days of life will affect a person’s health throughout life[33-34].It is known that maternally transfered microbiota plays a role in the establishment and maturation of infant intestinal microbiota.For example,the fetus is insulated in mother’s womb and is exposed to a direct environment where a microbiome has been detected both by culture-dependent and culture-independent methodologies[35].Environmental microbes rapidly colonize the neonatal gut soon after birth.The method of delivery method plays an important role in the initial establishment of infant gut microbes[36].Arboleya et al.[37]proposed that vaginal delivery and exclusive reliance on breastfeeding in early life are the gold standard for the establishment and development of healthy infant gut microbiota.Bifidobacterium,Bacteroides,andClostridiumproliferate and become the dominant genera associated with early life[38].Compared with spontaneous labor,infants born by cesarean section lose their only chance of direct contact with mother’s vaginal microbiome.Instead,their first exposure is the environment and the skin of the mother.As a result,opportunistic pathogens from the hospital environment,such asEnterococcus,Enterobacter,andKlebsiella,were found in their colon[39].At the same time,the early colonization of intestinal microbiome of cesarean infants was similar to the skin of the mother,mainly composed of facultative anaerobe,includingStaphylococcus,Propionibacterium,Corynebacterium,BacteroidesandBifidobacteriumwith low relative abundance[40].Our results observed thatB.breveandB.pseudocatenulatumwere the dominant bacteria in the infant’s gut,even though the infant participated in this study was delivered by cesarean section.The high resemblance between the faecal microbiota composition of the subject of this study and those delivered vaginally could be a result of maternal Probio-M9 supplementation in late pregnancy.A previous study of Ismail et al.[41]found that providing mothers with LGG in late pregnancy could increase the amount of gut bifidobacteriain infants.Toscano et al.[42]found that oral intake ofL.rhamnosusHN001 regulated the host intestinal microbiota by reducing potentially harmful bacteria while increasing beneficial bacteria,such as bifidobacteria.These results are consistent with the current observation that Probio-M9 not only could be vertically transmitted from mother to infant via oral intake of the bacteria but also colonized in the infant gut during early life.
In this study,the infant didn’t have any other sources of food during lactation.This also made the infant’s digestive tract and gut microbiota less susceptible to other factors.Our data also showed that the gut microbiota composition of the infant was much less complicated than that of the mother,which is in line with findings of previous studies.Moreover,due to the relative simplicity of the infant gut microbiota structure,it is likely to be influenced by external factors,such as the environment,and both infant’s and mother’s diet[36].In contrast,the gut microbiota of healthy adults is more stable,which does not fluctuate as much with external factors like probiotic consumption.For example,Toscano et al.[42],found that the administration ofL.rhamnosusHN001 andB.longumBB536 did not affect the diversity and richness of intestinal microbiota in healthy people.These observations are also consistent with current findings.
By the richly differentiated taxonomic correlation network between the mother and infant,we speculated that Probio-M9 supplementation during pregnancy promoted the colonization ofB.brevein the intestine of infant,and inhibited the growth of a variety of bacteria other thanBifidobacteriumin the intestine of infant,helping the infant to build a relatively healthy intestinal environment at the beginning of life.
The composition and activity of the intestinal microbiota co-develop with the host from birth,and are influenced by complex interactions with the host genome,nutrition,and lifestyle.Intestinal microbiota is involved in the regulation of multiple host metabolic pathways,mediating interactions between the host and bacterial metabolism,signal transduction,and immune and inflammatory axes.These axes connect the gut,liver,muscle,and brain physiologically.An in-depth understanding of these axes is the prerequisite for optimizing therapeutic strategies to manipulate the gut microbiome to fight disease and improve health[43].Within these metabolic axes,multiple bacterial genomes could sequentially regulate metabolic responses,leading to combined metabolism of microbiome and host genomes to substrates[44].The gut microbiota communicates metabolically with the host in a coordinated manner.
A number of differential metabolic pathways were found between the mother and the infant,which could be classified into three categories.The first category of metabolic pathways only existed in infant’s data subset,most of which were related to basic life activities such as mitochondrial activity and biosynthesis.These included PWY-7269,PWY-7245,PWY-7268,PWY0-1241,PWY-6396,and so on.The third category of metabolic pathways existed mostly in maternal gut,most of which were related to glucose metabolism,amino acid synthesis,short chain fatty acids (SCFAs) production and so on.For example,PWY-5676 represented the acetyl-CoA fermentation to butanoate II pathway,which regulates the synthesis of butyric acid.Acetic acid,propionic acid,and butyric acid are the main SCFAs in the intestine and are the most important products of the intestinal microorganisms[45].As the main energy source of intestinal cells,SCFAs regulate the absorption of a variety of nutrients in the intestinal tract and are widely involved in energy metabolism.This category of pathways is rather adult-specific.Our results showed that there were significant differences in the gut microbiota environment and metagenomic potential between infants and adults.
In conclusion,we found that maternal intake of Probio-M9 during the final weeks of gestation could deliver these bacteria to the infant vertically,and the bacteria could colonize the infant intestine until the end of the trial period.The infant gut was found to be colonized by a high proportion of bifidobacteria,which might have been a positive effect related to maternal Probio-M9 intake.One major limitation of this work was that only one volunteer participated in this study.Although this sample has no statistical significance and is not a traditional cross-sectional study,it is a long-term continuous follow-up study on the sample,thus discovering the above rules,and reporting and revealing some phenomena for the first time that Probio-M9 may be able to deliver vertically from mother to infant and so on.In the subsequent research,we will conduct animal experiments or large-scale human trials to verify and decipher the current findings and their mechanism.Meanwhile,these findings also guide the next experimental direction for our research.
Conflict of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by Science and Technology Major Projects of Inner Mongolia Autonomous Region (2021ZD0014).
- 食品科学与人类健康(英文)的其它文章
- Protective effects of oleic acid and polyphenols in extra virgin olive oil on cardiovascular diseases
- Diet and physical activity inf luence the composition of gut microbiota,benef it on Alzheimer’s disease
- Inf luence of nitrogen status on fermentation performances ofnon-Saccharomyces yeasts: a review
- Ganoderma lucidum: a comprehensive review of phytochemistry,eff icacy,safety and clinical study
- Resveratrol combats chronic diseases through enhancing mitochondrial quality
- Demonstration of safety characteristics and effects on gut microbiota of Lactobacillus gasseri HMV18

