Characteristics of Mycobacterium tuberculosis serine protease Rv1043c in enzymology and pathogenicity in mice
TANG Yang-yang, CUl Ying-ying, JlANG Yan-yan, SHAO Ming-zhu, ZANG Xin-xin, DANG Guang-hui,LlU Si-guo
State Key Laboratory for Animal Disease Control and Prevention, Harbin Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Harbin 150069, P.R.China
Abstract The serine proteases of Mycobacteria tuberculosis (Mtb) are important contributors to the process of bacterial invasion and its pathogenesis.In the present study, we systematically characterized the role of the Rv1043c protein in Mycobacterium infection by purifying the Rv1043c protein in Escherichia coli and constructing a Mycobacterium smegmatis (Msg) strain overexpressing Rv1043c (Msg_Rv1043c).We found that Rv1043c had serine protease activity and localized to the surface of Mtb.We determined that the optimal pH and temperature for the Rv1043c serine protease were 9.0 and 45°C, respectively.Moreover, the serine protease activity of Rv1043c was enhanced by divalent metal ions of Ca2+ and Mg2+.Site-directed mutagenesis studies demonstrated that the serine 279 residue in Rv1043c plays a catalytic role.Additionally, mouse model studies confirmed that Rv1043c significantly enhanced the survival of Msg in vivo, induced pulmonary injury and lung cell apoptosis, and promoted the release of pro-inflammatory cytokines interleukin-1β and interleukin-6 in mice.This study presents novel insights into the relationship between mycobacterial serine protease and the pathogenesis of the disease.
Keywords: Mycobacterium tuberculosis, Mycobacterium smegmatis, serine protease, Rv1043c, pathogenicity
1.lntroduction
Mycobacteriumtuberculosis(Mtb) is the pathogen that causes tuberculosis (TB), which is a major public health problem.Approximately 1.28 million deaths and 9.87 million new cases of TB were reported globally (Rahlweset al.2023).Infections by multidrug-resistant Mtb and human immunodeficiency virus (HIV) pose major challenges to TB control (Diedrichet al.2016).It has been reported that Mtb, as an intracellular bacterium,can persist and replicate within alveolar macrophages after ingestion, leading to an asymptomatic state in TB patients (Gomez and Mckinney 2004; Pieters 2008).Understanding the mechanism that enables Mtb’s persistence and self-protection against the host immune defenses is important.
Serine protease in bacteria has a pivotal role in the invasion of target cells and immunomodulation (Hedstrom 2002).The IgA1-specific serine protease ofNeisseria meningitidescan degrade IgG3 while promoting virulence(Spoerryet al.2021).Vibriocholeraeserine protease was shown to cleave intelectin, which suppresses the interaction between intelectin andV.cholera(Howellet al.2019).Mtb serine protease is well established to have pathogen-associated elements, while Rv3194c inhibits macrophage migration and enhances the intracellular persistence in the lungs (Liet al.2019).Evidence has shown that Rv3668c can affect host cytokine expression and apoptosis through Erk-NF-κB (Zhaoet al.2014).Serine protease HtrA contributes to virulence by cleaving the tight junction component claudin-8 inCampylobacter jejuni(Sharafutdinovet al.2020).Thus, virulent Mtb manipulates serine protease to adapt to macrophages’immune attack and survive.
The mode by which the host reacts to infection by Mtb relies on the critical role of macrophages, and diverse cellular activities can be detected after Mtb evasion,including necrosis, autophagy, and apoptosis (Tamgueet al.2019; Chaiet al.2020; Parket al.2021).Apoptosis is a mechanism by which macrophages act against pathogens that is proposed to limit bacterial growthin vivo(Bahet al.2020).Several reports have indicated that virulent strains mainly induced necrosis although apoptosis is a common response identified among macrophages (Chenet al.2006; Beharet al.2011).The different immune responses of the host during Mtb infection are affected by many virulence factors.
In particular, apoptosis inhibition is generally considered a single pattern by which Mtb exhibits defective antigen presentation (Liuet al.2015).The reported factors that suppress apoptosis include PtpA,Rv3354, NuoG, and SodA (Velmuruganet al.2007; Jainet al.2011; Danelishviliet al.2014; Wanget al.2017).Well-known antigens that enhance apoptosis include Rv0183, Rv0901, ESAT-6, and OppD (Samtenet al.2009;Zhanget al.2009; Xuet al.2010).Furthermore, genes involved in apoptosis are causes for concern since they help bacteria to adapt to their environment.
Here, we report the capacity of enzyme activity of serine protease Rv1043c in Mtb, and its detailed pathogenic roleinvitroandinvivo.We firstly identified Rv1043c as a new serine protease of Mtb, a recombinant Msg strain defined as Msg_Rv1043c was constructed, and we evaluated the pathogenicity of Rv1043c or its effect on macrophage apoptosis.We found that Rv1043c can promote the intracellular survival of Msg and cytokines release, which involves Mtb infection and pathogenesis.
2.Materials and methods
2.1.Bacterial strains and plasmid
Msg mc2155 and the recombinant strains Msg_pAIN,Msg_Rv1043c, and Msg_Rv1043cS279Awere grown in Middlebrook 7H9 medium (BD Biosciences, San Jose,CA, USA) and supplemented with 0.05% Tween 80(Amresco, Solon, OH, USA) and 0.2% glycerol (Sigma-Aldrich, St.Louis, MO, USA) with or without kanamycin(Prokesovaet al.1992).pAIN shuttle vector and pET-28a plasmid were stored in our laboratory.
2.2.Expression, protein purification, and preparation of rabbit polyclonal antibody
Therv1043cgene (Appendix A) was amplified from the genome of Mtb H37Rv, followed by cloning into the pET-28a.TheEscherichiacoliBL21 (DE3) strain was adapted for recombinant protein expression by the addition of isopropyl-β-D-thiogalactopyranoside (IPTG).The expressed bacteria were collected, and cell pellets were resuspended in 20 mmol L-1of Tris-HCl, 150 mmol L-1of NaCl, and 5% glycerol (pH 8.0), then disrupted by ultrasonication.Next, the precipitate was collected by centrifugation and resuspended in 20 mmol L-1of Tris-HCl,150 mmol L-1of NaCl, 0.5% SKL, and 5% glycerol (pH 8.0).The dissolved supernatant was dialyzed in 20 mmol L-1of Tris-HCl, 150 mmol L-1of NaCl, and 10% glycerol(pH 8.0).After 24 h, the dialyzed protein was clarified by centrifugation, and the supernatants were loaded onto a Ni Sepharose 6 fast flow resin (GE Healthcare,Chicago, IL, USA).The useless proteins were washed with 20 mmol L-1of Tris-HCl, 150 mmol L-1of NaCl, 5%glycerol, and 20, 40, and 60 mmol L-1of imidazole (pH 8.0).The recombinant Rv1043c proteins (rRv1043c) were analyzed by SDS-polyacrylamide gel electrophoresis(SDS-PAGE).The purified protein was centrifuged in a 10-kDa Centricon concentrator (Merck Millipore,Burlington, MA, USA) to be replaced with 20 mmol L-1of Tris-HCl (pH 8.0).A total of 5 mg of rRv1043c protein was then used for preparation and antigen affinity purification of anti-Rv1043c rabbit polyclonal antibody by Biodragon Immunotechnologies.
2.3.Determination of the Rv1043c serine protease activity
To determine the Rv1043c serine protease activity, 10 μg of purified rRv1043c protein was incubated with 4 μg of casein (Sigma-Aldrich) in 20 μL of reaction buffer(20 mmol L-1of Tris-HCl; pH 8.0) at 37°C for 12 h.The mixture was analyzed by 10% SDS-PAGE.To quantify the Rv1043c serine protease activity, the assay was performed using a protease fluorescent detection kit (Sigma-Aldrich) according to the manufacturer’s instructions.The effects of divalent metal ions on the Rv1043c serine protease activity were determined by adding 5 mmol L-1of different divalent cation salts (BaCl2,CaCl2, MgCl2, ZnCl2, CuCl2, FeCl2, NiCl2, or MnCl2) as shown in Appendix B into the reaction at 45°C (pH 9.0)for 12 h.The results were analyzed at 280 nm using a multimode microplate reader.The experiments were conducted in triplicate.
2.4.Determination of optimal temperature, pH, and kinetic constant of Rv1043c
The influence of temperature on the Rv1043c serine protease was measured in 20 mmol L-1of Tris-HCl (pH 8.0) over a temperature range of 31-49°C (at intervals of 2°C).The impact of pH on the Rv1043c serine protease activity was determined under an optimal temperature in 40 mmol L-1of Britton-Robinson buffer solution (pH 3.0-12.0; at intervals of 1.0).The activity of Rv1043c was quantified using a previously described method(Liet al.2019).Briefly, 200 μL of reaction mixtures containing 40 μg of casein and 20 μg of rRv1043c protein was incubated at 37°C for 12 h.One enzymatic activity unit was defined as the amount of enzyme required to increase absorbance at 280 nm of 0.001 by mg-1h-1cm-1at 45°C.For Michaelis-Menton kinetic parameter determination, 20 μg of rRv1043c protein was incubated with 10 μL of different concentrations of casein (0.0125,0.025, 0.05, 0.1, 0.2, 0.4, 0.6, 0.8, or 1.6 mg mL-1) at 45°C (pH 9.0) for 12 h.The rRv1043c protein was substituted with 40 mmol L-1of Britton-Robinson buffer solution (pH 9.0) as a negative control.All reactions were terminated with 50 μL of 50% TCA (w/v) and measured at 280 nm using the multimode microplate reader (EnSpire;PerkinElmer, Waltham, MA, USA).All assays were repeated three times.
2.5.Site-directed mutagenesis
The pET28a-H160A, pET28a-D199A, and pET28a-S279A were constructed with reference to a previously reported method (Danget al.2018).Briefly, the amplification of recombinant mutant plasmids was carried out using complementary mutagenic oligonucleotide pairs(Appendix C) andrv1043c-pET-28aas a template.After identification, they were transformed into BL21 (DE3)cells and corresponding mutant proteins were obtained according to the expression conditions of rRv1043c protein.The mutant’s H160A, D199A, and S279A proteins were expressed and purified; then, the serine protease concentration was determined and compared to the amount of the wild-type rRv1043c protein using the aforementioned methods.
2.6.Circular dichroism spectroscopy analysis
Circular dichroism (CD) spectroscopy analysis was performed using the purified rRv1043c, rH160A, rD199A,and rS279A proteins (0.1 mg mL-1) dissolved in 20 mmol L-1of Tris-HCl (pH 8.0).The rRv1043c protein was detected over a range of pH values (3.0-11.0 at intervals of 1.0) after being dissolved in Britton-Robinson at room temperature (25°C) and over a range of temperatures(20-70°C at intervals of 10°C) after being dissolved in 20 mmol L-1of Tris-HCl (pH 8.0).Additionally, the rRv1043c, rH160A, rD199A, and rS279A proteins were dissolved in 20 mmol L-1of Tris-HCl (pH 8.0) and detected at room temperature.The UV CD spectra were collected in the wavelength range of 190-260 nm using the MOS450 circular dichroism analyzer (BioTools, Jupiter,FL, USA).All the experiments were repeated three times.
2.7.Construction of the recombinant strains and expression of rv1043c in Msg
Therv1043cgene from Mtb H37Rv was amplified using specific primers (listed in Appendix D).Therv1043cPCR products and the pAIN vector were combined using homologous recombinant.The recombinant plasmid pAIN-Rv1043c or empty pAIN vector was electroporated into Msg.The Msg harboring therv1043cgene was labeled as Msg_Rv1043c, while the harboring pAIN vector was labeled as Msg_pAIN.Msg_Rv1043c and Msg_pAIN strains were cultured in Middlebrook 7H9 broth to an OD600of 0.6-0.8.Then, the recombinant protein was induced with 0.4% acetamide for 24 h.Aliquots of induced mycobacterium strains were prepared in Middlebrook 7H9 broth supplemented with 20% glycerol and preserved at -80°C.The CFU per mL of the stored strains was quantified by plating serial dilutions on LB agar and determination of OD600.These stored strains were used for all subsequent infections of murine peritoneal macrophages and C57BL/6J mice.
2.8.Localization of the Rv1043c protein in recombinant Msg and Mtb
Preparation of subcellular fractions of recombinant Msg_Rv1043c and Mtb H37Ra was executed as described in previous reports from our laboratory (Dangetal.2018).The compositions of whole-cell lysate proteins(CLs), culture filtrate (CF), cell wall (CW), cytoplasm(CP), and cell membrane (CM) were separated by 10%SDS-PAGE and then transferred onto a polyvinylidene fluoride membrane (PVDF) (0.2 μm, Merck Millipore).The primary antibodies used included rabbit anti-Rv1043c(1:1 000 in PBST), mouse anti-Ag85b (1:500 in PBST;Abcam, Cambridge, UK), and rabbit anti-KatG (1:1 000 in PBST).Dylight680-conjugated anti-mouse or Dylight800-conjugated anti-rabbit IgG antibody (1:10 000;KPL, Gaithersburg, MD, USA) was used as a secondary antibody.Finally, the membrane was visualized and analyzed using the Li-Cor Odyssey imaging system (Li-COR Biosciences, Lincoln, NE, USA).
Mtb H37Ra was grown in 7H9 medium supplemented with 0.2% glycerol, 0.05% Tween 80, and 10% OADC to an OD600of 0.8-1.0.Then, after being collected by centrifugation at 5 000 r min-1for 5 min and washed with PBS, the precipitation was fixed with fixative for immune electron microscopy (Servicebio, Wuhan, China) at 4°C for 2 h.The samples were wrapped in 2% agarose,dehydrated in an ascending series of ethanol (30-100%),penetrated, and embedded in resin.We obtained 70-80-nm ultrathin sections using a Leica UC7 ultra-microtome and collected 150-mesh nickel grids with formvar film.Then we blocked them in 1% BSA for 30 min at room temperature, incubating first with rabbit anti-Rv1043c antibody (1:200 in 1% BSA) overnight at 4°C and then with a 10-nm gold-conjugated goat anti-rabbit secondary antibody (1:80 in 1% BSA; Sigma-Aldrich).Finally,ultrathin cryosections were stained with a 2% uranium acetate-saturated alcohol solution under light avoidance for 8 min.Samples were examined with the HT7800 transmission electron microscope.
2.9.Preparation of murine peritoneal macrophages
Murine peritoneal macrophages were collected from 6-to 8-week-old mice intraperitoneally injected with sodium thioglycolate medium (MERCK, Darmstadt, Germany).After centrifugation, peritoneal macrophages were cultured in RPMI 1640 supplemented with 10% (v/v) FBS(Gibco Laboratories, Gaithersburg, MD, USA).After 24 h of culture, the non-adherent cells were removed, and the obtained cells were confirmed to be murine peritoneal macrophages.
2.10.LDH measurement
Murine peritoneal macrophages were cultivated and treated with Msg_Rv1043c, Msg_Rv1043cS279A, and Msg_pAIN (multiplicity of infection=5).The supernatants were collected using a CytoTox 96 non-radioactive cytotoxicity assay (Promega Corporation, Madison, WI, USA) to measure the LDH release according to the manufacturer’s protocol after 12, 24, 36, and 48 h of infection.
2.11.Mouse infection, colony-forming unit (CFU)counting, and cytokine measurement
The C57BL/6 mice (n=5 per group) were intravenously infected with the Msg_pAIN, Msg_Rv1043c, and Msg_Rv1043cS279Arecombinant strains in 100 μL of PBS with 0.05% Tween 80 (PBST) at 5×107CFUs per mouse.Control mice were treated with PBST.At 1, 7, 14, 21,and 28 days after infection, the mice were euthanized.The bacterial loads in the lungs of infected mice were assessed as previously described (Danget al.2015).The serum cytokine production was examined with a commercially available mouse interleukin (IL-6 and IL-1β), tumor necrosis factor (TNF-α) ELISA Kit (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions.
2.12.Gross and histopathological analyses
At 14 days after infection, the left lungs were removed and photographed following euthanasia, and then fixed in 4% paraformaldehyde for tissue section and staining with hematoxylin and eosin (H&E) staining.The gross pathological score was assigned to the lungs based on the percentage and severity of the lung surface hemorrhage.Specifically, the percentage of lung hemorrhage was scored as follows: 0=no area, 1≤10%,2=10-20%, 3≥20-30%, 4≥30-40%, and 5≥40% of the lung surface.Meanwhile, the severity of lung hemorrhage was scored from 0-4 points, where 0=absent, 1=mild,2=moderate, 3=marked, and 4=severe.Separately, an overall histopathology score was assigned to the lungs based on the extent of inflammatory cell infiltration and alveolar septum widening of the lung sections.The percentage of inflammatory cell infiltration was scored from 0-5 points, where 0=no area, 0=no area, 1≤10%,2=10-20%, 3≥20-30%; 4≥30-40%, and 5≥40% of the lung section.Furthermore, the extent of alveolar septum widening was scored from 0-4 points, where 0=absent,1=mild, 2=moderate, 3=marked, and 4=severe.
2.13.TUNEL apoptosis analyses
Paraffin sections of mouse lungs obtained above were treated by deparaffinization and rehydration, antigen retrieval, permeabilization, and blocking with endogenous peroxidase.Then, the tissue samples were stained using a TUNEL Assay Kit (Servicebio, Wuhan, China) according to the manufacturer’s instructions.The apoptosis rate was quantified by calculating the number of apoptotic cells under a 200× field of vision in three to five tissue sections.
2.14.Statistical analysis
We performed all experiments at least three times in biological triplicate.Means and standard errors of means(SEM) are presented.The data were analyzed with the GraphPad Prism (GraphPad Software, San Diego, CA,USA) using one-way or two-way analysis of variance.Significance was denoted as follows:*,P<0.05;**,P<0.01;***,P<0.001; or****,P<0.0001.
3.Results
3.1.Rv1043c has protease activity and is a cell surface-associated protein of Mtb
The Mtbrv1043cgene encodes a serine protease of 36.1 kDa.Multiple sequence alignments showed that Rv1043c is highly conserved among pathogenic mycobacteria and non-existent in non-pathogenic mycobacteria like Msg (Fig.1) which indicated that Rv1043c might be a virulence factor.
To study the characteristics of the Rv1043c protein,which was expressed inE.coliBL21 (Fig.2-A and B) and purified using a Ni-NTA chelating affinity chromatography(Fig.2-C), the purified Rv1043c protein was co-incubated with FITC-labeled casein.This analysis revealed that the Rv1043c protein could cleave the FITC-labeled casein(Fig.2-D) and casein (Fig.2-E) substrates.To determine the subcellular localization of Rv1043c, we performed a Western blot assay of Mtb H37Ra and Msg_Rv1043c cell fractionation and found that Rv1043c was measured largely in the cell wall fractions of Mtb H37Ra and Msg_Rv1043c (Fig.2-F and G).As expected, the control of Ag85B was largely present in the cell wall and culture filtrate, while KatG was found only in the cytoplasmic fraction (Fig.3-F and G).Moreover, immuno-electron microscopy further verified the unique localization of Rv1043c at the surface of Mtb H37Ra (Fig.3-H).These results showed that Rv1043c is an extracellular protease and might play a role in the interaction with the host.
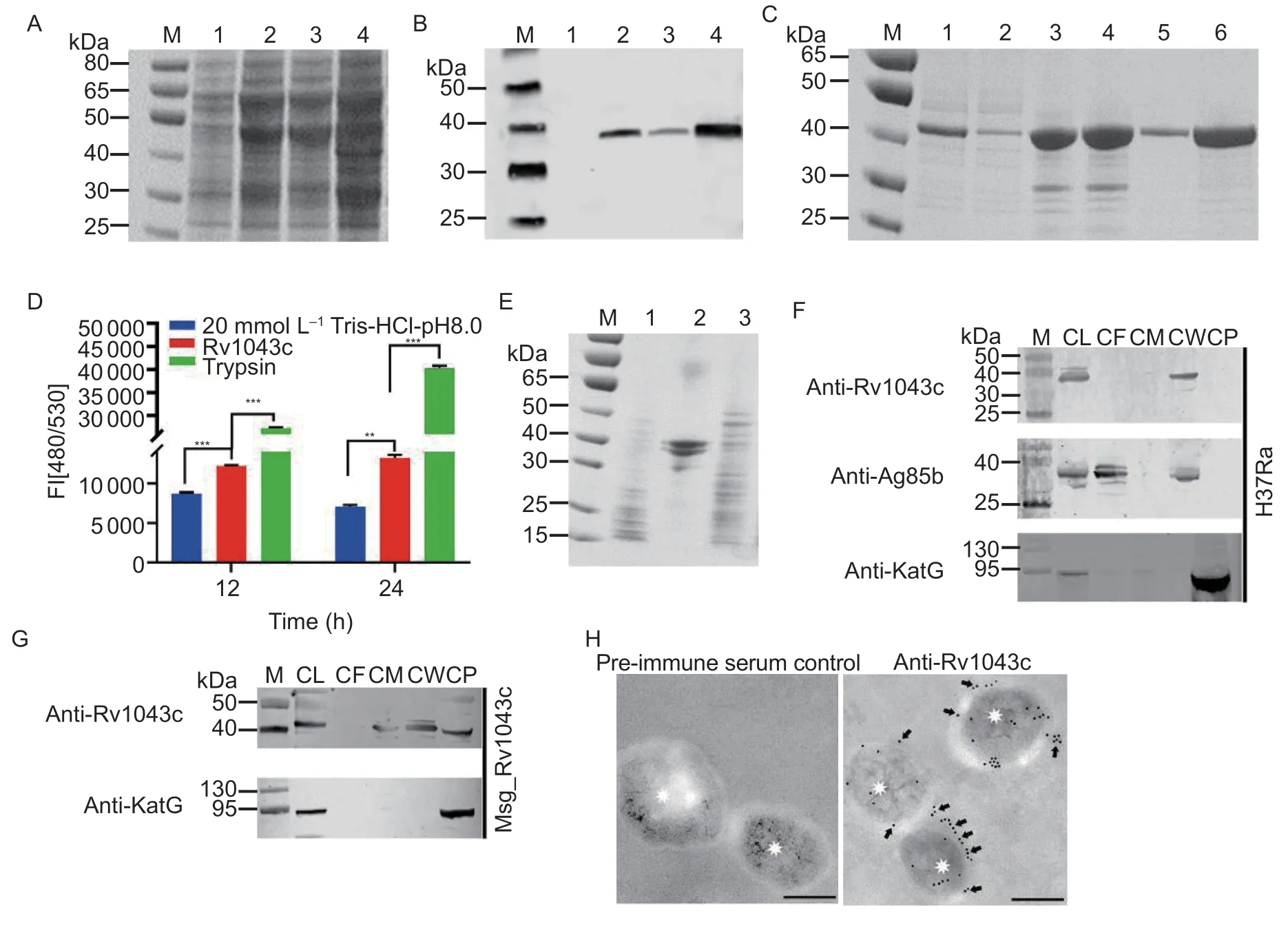
Fig.2 Rv1043c has serine protease activity and expresses on the mycobacterial surface.A and B, SDS-PAGE (A) and Western blot (B) analysis of the expression of the Rv1043c protein.Lane 1, Rv1043c protein before IPTG induction; lane 2, Rv1043c after IPTG induction; lane 3, supernatant of Rv1043c after induction; lane 4, precipitation of Rv1043c after induction.C, SDSPAGE analysis of the affinity chromatography purification of the Rv1043c protein.Lane 1, Rv1043c protein after renaturation;lane 2, passing-through from the resin; lane 3, protein-bound resin before washing; lane 4, protein-bound resin after washing;lane 5, purified Rv1043c protein eluted by 250 mmol L-1 imidazole; lane 6, Rv1043c protein eluted by 250 mmol L-1 imidazole.D,fluorescent quantitative analysis of the Rv1043c serine protease activity with FITC-casein substrate.The results were analyzed by GraphPad Prism using two-way ANOVA followed by Bonferroni’s multiple comparison post hoc test.Data are expressed as mean±SE (n=3).Asterisks represent a significant difference (**, P<0.01; ***, P<0.001).E, SDS-PAGE analysis of the Rv1043c serine protease activity with casein substrate.Lane 1, Rv1043c protein; lane 2, casein; lane 3, co-incubation of casein and Rv1043c.F and G, immunoblot analysis of whole-cell lysates (CLs), culture filtrate (CF), cell membrane (CM), cell wall (CW), and cytoplasm(CP) collected by ultracentrifugation of Mtb H37Ra and Msg_Rv1043c.Ag85B (secreted and cell wall) and KatG (cytoplasm) were used as fractionation controls.H, immuno-electron microscope analysis for localization of Rv1043c labeled with 10-nm gold (black arrows) on ultrathin sections of Mtb H37Ra.Staining controls for anti-Rv1043c were performed with pre-immune serum.Asterisks indicate the Mtb H37Ra cells.Scale bars=200 nm.
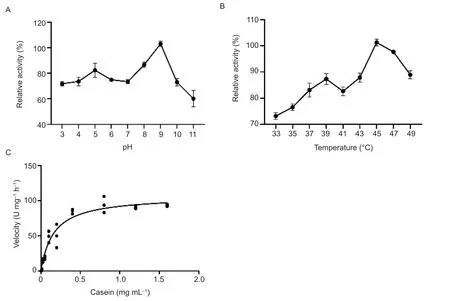
Fig.3 The characteristics of the Rv1043c serine protease.A,effects of pH on the Rv1043c protease activity.B, effects of temperature on the Rv1043c protease activity.C, Michaelis-Menton kinetic of the Rv1043c protease activity.The reaction was subjected to 40 mmol L-1 of Britton-Robinson (pH 9.0)and 5 mmol L-1 of CaCl2 for 12 h at 45°C.Km values for casein are (0.1±0.002) mg mL-1, and Vmax values for serine protease activity are 91.29 U mg-1 h-1.The errors are represented as the SE of three independent experiments.
3.2.Effects of pH, temperature, and divalent cations on the Rv1043c serine protease
Measurements of the Rv1043c serine protease activity across a wide pH and temperature range revealed that the optimal pH and temperature for Rv1043c were 9.0 and 45°C, respectively.The effect of different divalent cations on the serine protease activity of Rv1043c was examined at 45°C and a pH of 9.0.The serine protease activity of Rv1043c was increased in the presence of CaCl2((125±12.67)%) or MgCl2((112±14.82)%) compared to in the absence of divalent cations (Appendix B).Other divalent cation salts (ZnCl2, CuCl2, FeCl2, NiCl2,and MnCl2) displayed different inhibitory effects of the Rv1043c serine protease activity (Appendix B).The kinetic parameters of the Rv1043c serine protease were evaluated using a casein substrate at 45°C (pH 9.0) in the presence of 5 mmol L-1of CaCl2.Michaelis-Menton kinetic assays showed that the Km value toward casein was (0.1±0.002) mg mL-1(Fig.3-C).The Vmaxvalue was 91.29 U mg-1h-1for casein activity (Fig.3-C).
3.3.Site-directed mutagenesis
To determine the active site on the Rv1043c serine protease, multiple sequence alignments were performed with reported serine protease-conserved domains using the MEGA5 Software (Fig.4-A).We replaced 160 histidine, 199 aspartic acid, and 279 serine residues with alanine, and the H160A, D199A, and S279A mutant proteins were produced by affinity chromatography.The H160A, D199A, and S279A mutant proteins caused 7,50.2, and 70% losses of activity, respectively, compared to wild-type Rv1043c (Fig.4-B and C).These results showed that the active site of Rv1043c is 279 serine residues.

Fig.4 The substitution of S-279 by alanine reduces the serine protease activity of the Rv1043c protein.A, multi-sequence alignment analysis of the Rv1043c protein sequence from the conserved domains of other reported bacterial serine proteases using the MEGA5 software and BoxShade server.Micromonas pusilla EEH52569.1 (CCMP1545), Stigmatella aurantiaca DW4/3-1(Q08Z65), and Streptomyces vitaminophilus MarP family serine protease (WP_018386153.1), Catenulispora acidiphila MarP family serine protease (WP_015797397.1), Mtb MarP (WP_011799367.1), Staphylococcus aureus subsp.aureus JH9 Serine protease SplC (A5ITX6.1), Escherichia coli K-12 protease degS (3LGV_C), and Bacillus subtilis subsp.subtilis str.168 serine protease YyxA(P39668.2).B and C, the serine protease activity of mutant proteins (H160A, D199A, and S279A) was analyzed by SDS-PAGE (B),lane 1, casein; lane 2, Rv1043c protein; lane 3, co-incubation of H160A and casein; lane 4, co-incubation of S279A and Casein,and ultraviolet spectrophotometry (C).Data are expressed as mean±SE (n=3).
3.4.Circular dichroism spectroscopy analysis of Rv1043c, H160A, D199A, and S279A
To determine the secondary structural stability of the Rv1043c protein, CD spectroscopy was conducted over the wavelength range from 190-260 nm.CD spectra data were collected over a range of pH values (3.0-11.0)at room temperature (25°C).The thermal stability of the Rv1043c protein were gathered over a range of temperatures from 20-70°C at a pH of 8.0 as shown in Fig.5-A.The curves converged at the pH values of 3.0,4.0, and 7.0-11.0 but were distorted at those of 5.0 and 6.0(Fig.5-B).We further analyzed whether the secondary structural stability of the mutant proteins H160A, D199A,and S279A.The conformation of the Rv1043c protein was stable with the curves converging (Fig.5-C).Interestingly, mutations at single-nucleotide sites did not cause significant structural changes, and there was almost no significant difference in the secondary structure between the mutant and Rv1043c.The results confirmed that the S279 amino acid residue significantly affected the activity of Rv1043c serine protease.
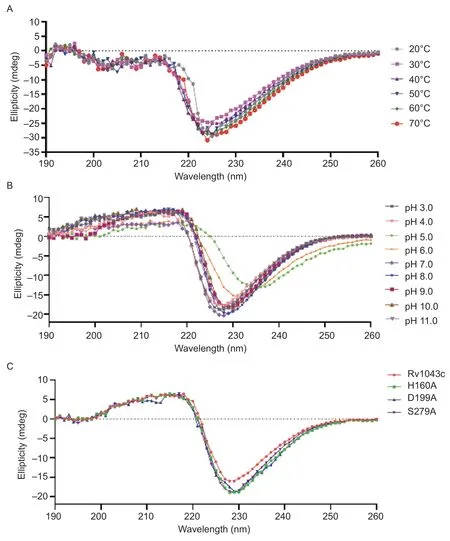
Fig.5 Stability analysis of the secondary structure of the wild-type Rv1043c and mutant (H160A, D199A, and S279A) proteins.A and B, the CD measurements of the Rv1043c protein were performed in buffers with pH values ranging from 3.0-11.0 at room temperature (A) and different temperatures ranging from 20-70°C at a pH of 8.0 (B).C, the CD measurements of Rv1043c, H160A,D199A, and S279A were conducted in 20 mmol L-1 of Tris-HCl (pH 8.0) at room temperature.
3.5.Rv1043c serine protease induces host cell apoptosis in vivo
Reports showed that Enterovirus 71 3C protease andStreptococcuspneumoniaeClpP protease induce host cell apoptosis (Leeet al.2013; Wenet al.2020).In Fig.6-A and B, TUNEL staining results and scoring showed that Rv1043c expression markedly increased the Msg infection-induced apoptosis in the lungs compared to Msg_pAIN and Msg_Rv1043cS279A.We examined whether Rv1043c serine protease induced host cell apoptosis.Mouse peritoneal macrophages infected with Msg_pAIN,Msg_Rv1043c, or Msg_Rv1043cS279Aas indicated were analyzed by LDH release assay.Our data showed that the expression of the Rv1043c serine protease in Msg caused strong cytotoxicity effects on mouse peritoneal macrophages compared to pAIN and Rv1043cS279A, as shown in Fig.6-C.The results confirmed that Rv1043c was a potential apoptosis- and virulence-related factor.
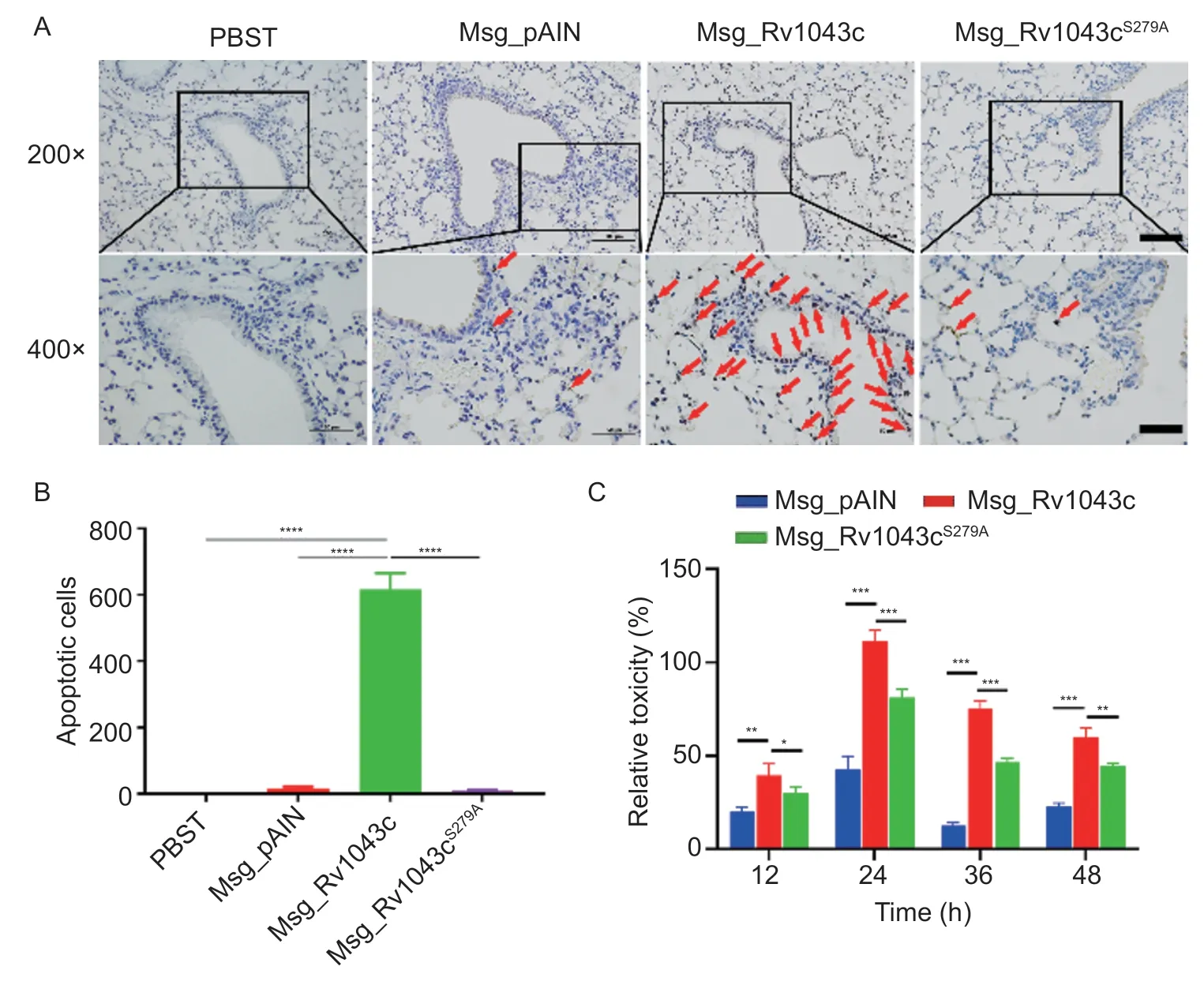
Fig.6 Rv1043c serine protease induces host cell apoptosis.A, TUNEL analysis in mice.The mice (n=5) were infected with Msg_pAIN, Msg_Rv1043c, or Msg_Rv1043cS279A as indicated; the red arrows indicate apoptotic cells.B, analysis of apoptotic cells.The apoptotic cells were counted in the selected visual field, and three parallel experiments were conducted.The asterisk indicates the significance: ****, P<0.0001.C, LDH analysis.The LDH release was measured in macrophages infected with Msg_pAIN,Msg_Rv1043c, or Msg_Rv1043cS279A.Data are expressed as mean±SE (n=3).*, P<0.05; **, P<0.01; ***, P<0.001.
3.6.Virulence of Rv1043c requires serine protease activity
To investigate the role of the Rv1043c serine protease in mycobacterium infection, we challenged C57BL/6 mice with Msg_pAIN, Msg_Rv1043c, or Msg_Rv1043cS279Aand evaluated gross pathology, histopathology, and bacterial load in the lungs.Mice were also treated with PBST as the control.For gross pathology, infection with Msg_Rv1043c led to a large area of hemorrhagic necrosis on the surface of the lungs, but those infected with Msg_pAIN or Msg_Rv1043cS279Aand treated with PBST had no pathological injury (Fig.7-A).For histopathology,Msg_Rv1043c-infected mice showed a large amount of inflammatory cell infiltration and alveolar septum widening of the lungs.However, Msg_pAIN-infected mice displayed a small amount of local congestion, while Msg_Rv1043cS279A-infected and PBST-treated mice experienced no pathological changes (Fig.7-C).In addition, gross-pathological and histo-pathological scores of the Msg_Rv1043c-infected mice showed a significant difference in injury-causing abilities compared to Msg_pAIN-infected, Msg_Rv1043cS279A-infected, and PBSTtreated mice (Fig.7-B and D).The mycobacterial load in the lung tissue of mice infected with Msg_Rv1043c was much higher than those of mice infected with Msg_pAIN or Msg_Rv1043cS279Aat 7, 14, and 21 days after infection(Fig.8).Together, these results indicate that the Rv1043c serine protease is an essential factor for mycobacterium virulence.
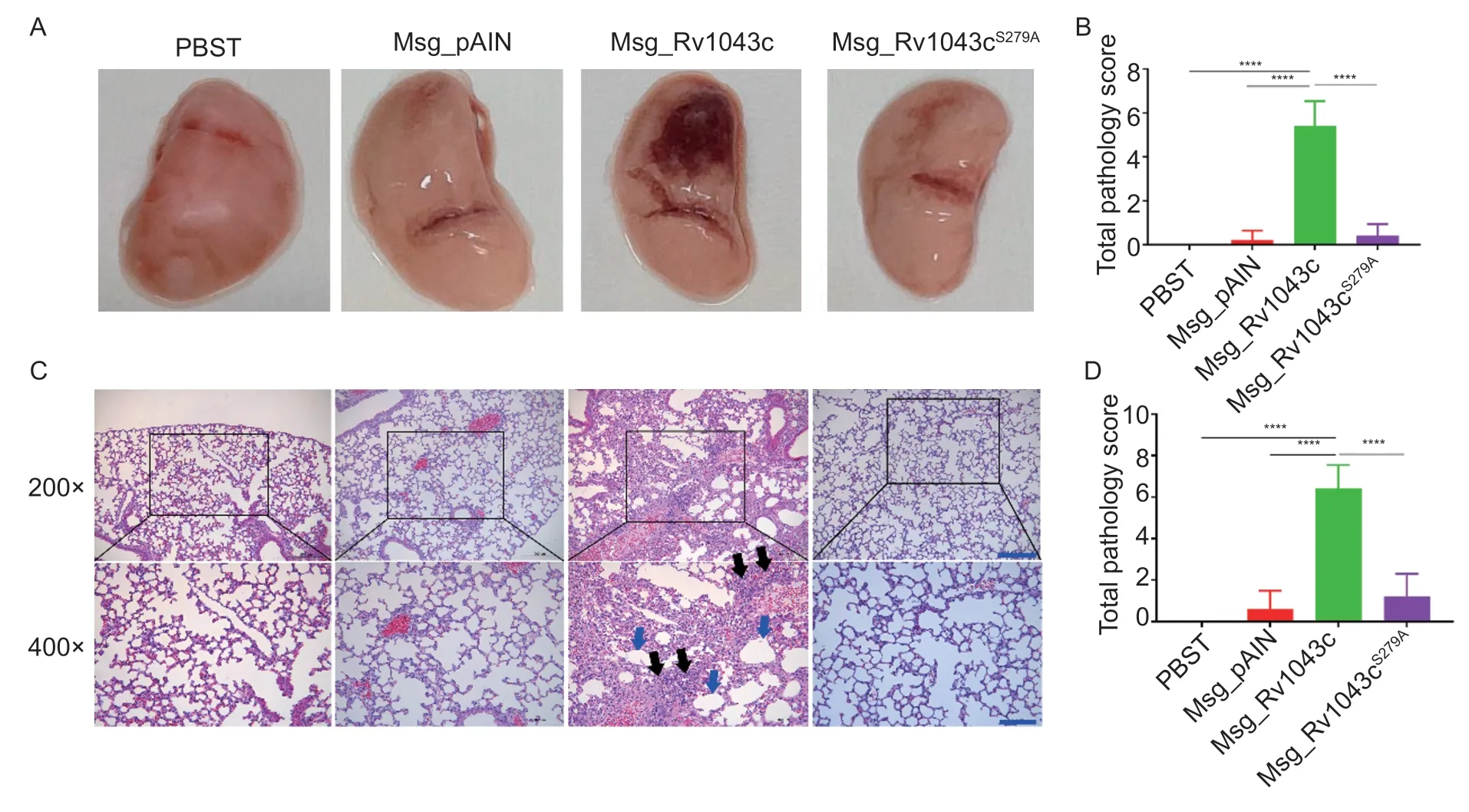
Fig.7 Rv1043c serine protease is an essential virulence factor for mycobacterium.C57BL/6 mice were intravenously infected with 5×107 CFUs per mouse of Msg_pAIN, Msg_Rv1043c, or Msg_Rv1043cS279A.Gross pathology (A) and histopathology (C)were evaluated in the lungs of mice infected for 14 days.The scores of lung pathology injury (B) from H&E-stained sections and histopathology injury (D) from H&E-stained sections were analyzed using Two-way ANOVA followed by Bonferroni’s multiple comparison post hoc test.Data are mean±SE (n=5).The asterisks represent the significance (****, P<0.0001).
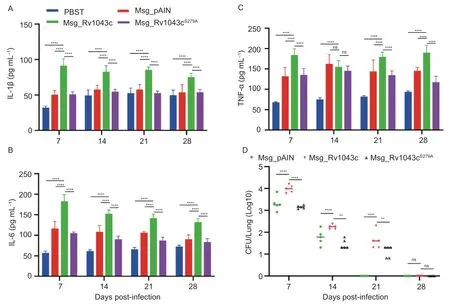
Fig.8 Expression of Mtb Rv1043c, but not Rv1043cS279A, in Msg induces increased production of IL-1β, IL-6, TNF-α and the persistence of mycobacteria in mice during infection.A-C, ELISA revealed that Mtb Rv1043c serine protease in Msg increases the levels of IL-1β (A), IL-6 (B), and TNF-α (C).The mice were infected or treated for 1, 7, 14, 21, and 28 days with Msg_pAIN,Msg_Rv1043c, Msg_Rv1043cS279A, and PBST.D, Mtb Rv1043c serine protease increases the persistence of Msg in mice infected.Data are showed as mean±SE (n=5).Two-way ANOVA with Bonferroni’s multiple comparison test was employed.**, P<0.01; ****,P<0.0001; ns, not significant.
3.7.Rv1043c serine protease upregulates the inflammation response and promotes mycobacterium persistence in mice
We further measured whether serine protease activity was essential for the Rv1043c-mediated promotion of inflammatory cytokine expression in mice during recombinant Msg infection.We found that the expression of Rv1043c in Msg upregulated the release of IL-1β and IL-6 in the sera of mice at 7, 14, 21, and 28 days post-infection, and that of TNF-α at 7, 21, and 28 days post-infection compared to the expression of pAIN or Rv1043cS279Ain Msg and PBST, respectively (Fig.8-A-C).Consistently, the expression of Rv1043c also promoted the persistence of mycobacteria in mice at 7, 14, and 21 days post-infection.However, Rv1043cS279Aserine protease mutation abolished the Rv1043c-mediated promotion of the persistence of mycobacterium (Fig.8-D).Taken together, these results indicate that the serine protease activity of the Rv1043c upregulates the release of inflammatory cytokines and promotes the persistence of mycobacteria in mice.
4.Discussion
Proteases, including serine protease, cysteine protease,aspartic protease, and other metalloproteinases, are widely distributed in bacteria and their hosts.Almost onethird of proteases are serine proteases, which participate in different diseases, including cancer and emphysema(Hedstrom 2002; Harish and Uppuluri 2018; Qinet al.2022).There is evidence that serine protease acts in connection with the evasion of host immune surveillance,the adaption of nutrition deficiency, and penetration into tissue (Skorko-Gloneket al.2017; Burchacka and Sienczyk 2018).The role of serine proteases in promoting intracellular infection and persistence is an interesting topic, but this is rarely reported in Mtb.The current study identified Rv1043c as a novel serine protease involved in pro-inflammatory reactions and apoptotic responses within the host.
In many Gram-positive and Gram-negative bacteria,cysteine protease and serine protease can degrade the cellular components of the host.The cleavage process of polyproteins and the MHC-II-mediated antigen presentation of epithelial cells are considered to be related to cysteine protease (Liuet al.2004; Yan and Wu 2021).In contrast to cysteine protease, recent research progress has demonstrated that HtrA can degrade extracellular matrix proteins, including fibronectin, as well as cadherin, which contributes to cell adhesion inC.jejuni,Helicobacterpylor, andBacillusanthracis(Skorko-Gloneketal.2017; Boehmet al.2018; Sharafutdinovetal.2020;Berneggeret al.2021).Our study found that Rv1043c and its homologs only exist in pathogenic strains rather than non-pathogenic mycobacterium such as Msg.We proposed that the Mtb can manipulate the invasion and adhesion process through serine protease as a potential adaptive strategy.In H37Ra and Msg_Rv1043c strains,Rv1043c was detected in the cell wall, which is consistent with the localization of protease with the function of cutting cell components.
Specifically, temperature and pH can have a significant impact on enzyme activity.Rv1043c retained 60% of the enzyme activity after incubation at pH values of 3-11,demonstrating that Rv1043c is stable across a broad pH range, although the highest degradation efficiency was observed at a pH of 9.Even if the highest enzyme activity is exerted under alkaline conditions and high temperatures, we believe that stable activity is very beneficial for Mtb to adapt to adverse external stimuli.The substitution experiment suggested that, for the conservation of amino acid residues of Rv1043c, the two sites 199D and 279S are essential for serine protease activity.In particular, the mutation of S279A resulted in the loss of 70% of the enzyme activity.The catalytic domain covering the G277NSGGP sequence was similar to the identified consensus sequence GDSGGP in trypsinlike serine protease (Hedstrom 2002).The highly diverse trypsin-like serine proteases shared a commonβ-barrel,while, due to the different lengths of loops formed by various proteases, there were certain differences in functions.The 279S site is a recognized catalytic site and located in the reported “191 loop”.Metal ions such as Ca2+and Zn2+can regulate the function of protease,including glycosylation sites (Goettiget al.2019).The enzyme activity characteristics of Rv1043c are consistent with those previously reported, and serine protease activity is also indispensable in virulence and apoptosis.
Mtb, as a highly adaptable intracellular bacterium, can evade the immune response of its host by controlling cellular functions such as autophagy, apoptosis, or pyroptosis (Liuet al.2017).An extrinsic or intrinsic way of inducing apoptosis is possible; the extrinsic pathway is stimulated by extracellular ligands and mediated by TNF or Fas receptors, and the intrinsic pathway is activated by intracellular signaling.Evidence has shown that Mtb is a well-known and crafty enemy.Furthermore,serine proteases Rv3090 employed by Mtb promote the late apoptosis of macrophages during infection, which benefits intercellular diffusion (Cuiet al.2022).To some extent, it is beneficial for the spread of Mtb to turn on the“apoptosis switch” of the infected macrophages rather than cell necrosis.The capability of Rv1043c to induce macrophage apoptosis has been verified.
A variety of bacteria also produce extracellular proteases as an important component of their virulence.It is a common mechanism that the expression level of serine protease was significantly increased after the invasion,activating protease-cleaved cytoskeletal components in addition to immune molecules (Prokesovaet al.1992;Rodriguez-Cabreraet al.2010; Sunet al.2017; Spoerryet al.2021).Meanwhile, the overexpression of Rv1043c induced a significant accumulation of IL-1β and IL-6,leading to severe pathological damage.IL-1β is mainly secreted by activated macrophages as an inflammatory mediator to protect the host from Mtb infection.IL-6 is a multifunctional pro-inflammatory cytokine; many reports have offered contradictory conclusions regarding IL-6’s role in Mtb infections (Ladelet al.1997; Sodenkampet al.2012).The generation of IL-6 in macrophages infected with Mtb is accountable for the suppression of Th1 response and IFN-γ-mediated autophagy (Shinet al.2010).Thus, we showed that Rv1043c participates in the immune response and virulence, which cannot be ignored.
5.Conclusion
In summary, we define Rv1043c as a novel serine protease involved in the pathogenesis and inflammatory reaction of Mtb infection.The induced apoptosis process and persistence within mice largely depend on serine protease activity.Admittedly, further investigation is needed into the pathway by which Rv1043c promotes apoptosis, and it is possible that Rv1043c can be used as the target of serine protease inhibitors in the process of limiting the spread of TB.
Acknowledgements
This research was supported by the National Key Research and Development Program of China(2021YFD1800403) and the National Natural Science Foundation of China (32273005 and 32002256).
Declaration of competing interest
The authors declare that they have no conflict of interest.
Ethical approval
This study was approved by the Animal Ethics Committee of Harbin Veterinary Research Institute, CAAS.All animal experiments followed the relevant provisions of animal welfare to alleviate pain (Ethical Committee Approval number HVRI-IACUC-210426-03).
Appendicesassociated with this paper are available on https://doi.org/10.1016/j.jia.2023.06.025
 Journal of Integrative Agriculture2023年12期
Journal of Integrative Agriculture2023年12期
- Journal of Integrative Agriculture的其它文章
- Biotechnology of α-linolenic acid in oilseed rape (Brassica napus)using FAD2 and FAD3 from chia (Salvia hispanica)
- Analyzing architectural diversity in maize plants using the skeletonimage-based method
- Derivation and validation of soil total and extractable cadmium criteria for safe vegetable production
- Effects of residual plastic film on crop yield and soil fertility in a dryland farming system
- Identifying the critical phosphorus balance for optimizing phosphorus input and regulating soil phosphorus effectiveness in a typical winter wheat-summer maize rotation system in North China
- Transcriptome-based analysis of key genes and pathways affecting the linoleic acid content in chickens
