6-Thioguanine incorporates into RNA and induces adenosine-to-inosine editing in acute lymphoblastic leukemia cells
Xue-Jio You,Lin Li,Tong-Tong Ji,Neng-Bin Xie,Bi-Feng Yun,*,Yu-Qi Feng
a Department of Chemistry,Sauvage Center for Molecular Sciences,Wuhan University,Wuhan 430072,China
b School of Public Health,Wuhan University,Wuhan 430071,China
c School of Pharmacy,Shanghai Jiao Tong University,Shanghai 200240,China
Keywords:6-Thioguanine RNA modification Inosine Leukemia Mass spectrometry
ABSTRACT 6-Thioguanine (6TG) is a widely used chemotherapeutic agent for the treatment of a variety of human diseases including acute lymphoblastic leukemia.After entry into cells,6TG is metabolically converted into 6-thioguanosine (SG) nucleotide that can be incorporated into the genome during DNA replication.SG in genomic DNA could induce cell death by triggering the post-replicative mismatch repair (MMR)pathway.Meanwhile,incorporation of 6TG into the CpG sites could perturb the global DNA methylation and gene regulation.However,the effect of 6TG on RNA modifications is still unknown.Adenosine-toinosine (A-to-I) editing in RNA is one of the most common post-transcriptional modifications in mammals and there is growing evidence showing the significant alteration of A-to-I RNA editing in tumor tissues compared to normal tissues.In the current study,we examined the incorporation of 6TG into RNA and investigated its effect on A-to-I editing of bladder cancer-associated protein (BLCAP) transcript in acute lymphoblastic leukemia cells.The results demonstrated that SG could be incorporated into various RNA species,with mRNA having the most abundant SG.In addition,the results showed 6TG treatment elevated A-to-I editing in BLCAP transcript through upregulating adenosine deaminase 2 acting on RNA (ADAR2),which eventually contributes to the decreased cell viability.This study highlights a new mechanism of the cytotoxicity of 6TG in inducing cell death.
6-Thioguanine (6TG) is among the most successful chemotherapeutic agents for treating a variety of human diseases including acute lymphoblastic leukemia (ALL) [1].After entry into cells,6TG is converted into 6-thioguanosine (SG) nucleotides that can be incorporated into genomic DNA through DNA replication [2].SG in genomic DNA can introduceSG:T base pair,which triggers the post-replicative mismatch repair (MMR) pathway and induces cell death [3].It was reported that theSG:T base pair could be recognized by mammalian MMR factors to a similar extent as G:T mispair [4–6].Cells deficient in MMR pathway exhibited significant increased resistance to 6TG treatment [7].
DNA epigenetic modifications have important functions and biological significance [8–14].Incorporation ofSG into genomic DNA could also affect DNA epigenetic modifications,such as 5-methylcytosine (5mC).Treatment of acute lymphoblastic leukemia cells (Jurkat-T) with 6TG resulted in the decreased level of 5mC in genomic DNA [15].The presence ofSG at the methylated CpG sites in DNA inhibited the methylation of its 5′ adjacent cytosine,whereasSG at the unmethylated CpG sites enhanced the methylation of the opposing cytosine in the complementary strand [15].In addition to DNA,6TG can be theoretically incorporated into RNA.Similar to DNA epigenetic modifications,the chemical modifications in RNA molecules can also delicately regulate numerous biological processes [16–27].However,how 6TG may affect RNA modifications is still unknown.
Adenosine-to-inosine (A-to-I) editing in RNA is one of the most common post-transcriptional modifications in eukaryotes [28].Ato-I editing is catalyzed by the adenosine deaminase that acts on RNA (ADAR) family of enzymes [28].Three ADARs have been identified in human.ADAR1 and ADAR2 are widely expressed and active enzymes [29].However,ADAR3 lacks detectable catalytic activity [29].A-to-I editing has the potential to change the RNA sequence since inosines can pair with cytosines and therefore they are recognized as guanosines by the ribosome during translation[30].If the A-to-I editing occurs in the coding regions,the sequence,structure and function of proteins can be altered [31].Thus,A-to-I editing plays important roles in various regulatory processes such as splicing [32],microRNA processing [33],microRNA targeting [34] and mRNA stability [35].Abnormal A-to-I editing may lead to various diseases [36],such as autoimmune[37],cardiovascular diseases [38],neurological diseases [39],and cancer development [40].
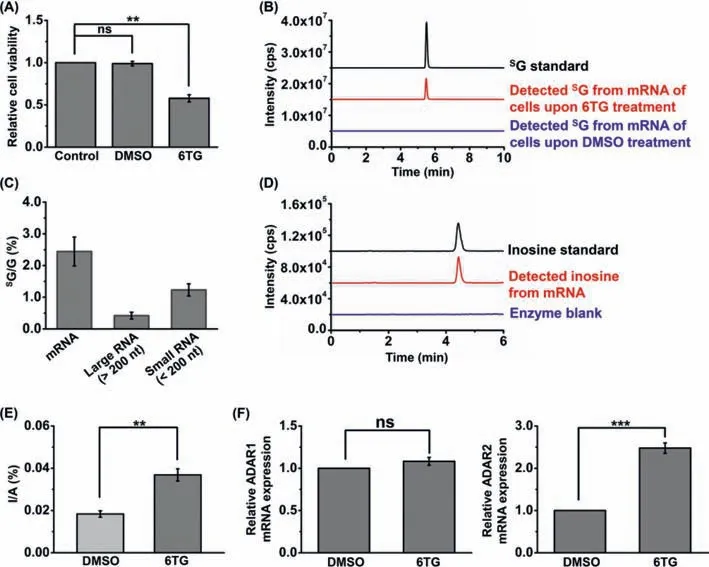
Fig.1.Evaluation of the incorporation of SG in RNA and investigation of the effect of 6TG on the level of inosine in mRNA.(A) 6TG treatment reduces cell viability.(B)Analysis of SG incorporated in mRNA by LC-ESI-MS/MS.Shown in black,red and blue are the extracted-ion chromatograms of SG standard,detected SG from mRNA of Jurkat-T cells upon 6TG treatment (3 μmol/L for 24 h),and upon DMSO treatment,respectively.(C) Quantification of SG incorporated in mRNA,large RNA (>200 nt) and small RNA (<200 nt) by LC-ESI-MS/MS analysis.(D) Detection of inosine in mRNA by LC-ESI-MS/MS analysis.Shown in black,red and blue are the extracted-ion chromatograms of inosine standard,detected inosine from mRNA of Jurkat-T cells,and enzyme blank control,respectively.Enzyme blank samples represent the samples only containing enzymes used for digesting RNA.(E) Quantification of inosine level in mRNA of Jurkat-T cells with and without 6TG treatment based on LC-ESI-MS/MS measurement.(F)The relative levels of ADAR1 and ADAR2 in Jurkat-T cells upon 6TG treatment (3 μmol/L for 24 h) compared to control (DMSO treatment).** P<0.01 (n=3); *** P<0.001(n=3); ns,not significant.
The accumulated lines of evidence show there is significant alteration of A-to-I editing in tumor tissues when compared with normal tissues [41].Bladder cancer-associated protein (BLCAP) was originally identified from human bladder carcinoma [42],which exerts tumor suppressor function in bladder cancer,tongue carcinoma,renal cancer,breast cancer and other malignant tumors[43,44].It has been reported that BLCAP transcript undergoes multiple A-to-I editing events [45] and the A-to-I editing in BLCAP transcript plays critical role in the development of cancers [46].Decreased level of A-to-I editing in BLCAP transcript was frequently observed in astrocytomas,bladder cancer and colorectal cancer when compared with the normal tissues [46].In the current study,we examined the incorporation ofSG into RNA and investigated its effect on A-to-I editing of BLCAP transcript in acute lymphoblastic leukemia cells.
We treated Jurkat-T cells with 6TG and examined the incorporatedSG in RNA.Firstly,it can be seen that treatment of human Jurkat-T cells with 3 μmol/L of 6TG for 24 h led to the decreased cell viability (Fig.1A).We then evaluated the incorporation of 6TG in different RNA species.To this end,the isolated mRNA,large RNA(>200 nt),and small RNA (<200 nt) from Jurkat-T cells upon 6TG treatment were enzymatically digested followed by LC-ESI-MS/MS analysis.The results showed thatSG could be distinctly detected in mRNA (Fig.1B),large RNA (>=200 nt) (Fig.S1A in Supporting information),and small RNA (<200 nt) (Fig.S1B in Supporting information).The quantification data showed that ~2.4%,0.4%,and 1.2% ofSG (SG/G) were found in mRNA,large RNA (>200 nt),and small RNA (<200 nt),respectively (Fig.1C).These results clearly displayed thatSG could be detected in different RNA species,with mRNA having the most abundantSG.We reason that mRNA has a faster turnover rate than other RNA species,which leads to the higher incorporatedSG in mRNA.
RNA modifications were discovered to play critical roles in regulating a variety of physiological processes [17].Here,we further investigated the effect of 6TG on modifications in mRNA.mRNA from Jurkat-T cells exposed to 6TG was isolated and enzymatically digested followed by LC-ESI-MS/MS analysis.The results showed that 13 modifications,includingN6-methyladenosine(m6A),N1-methyladenosine (m1A),N6,2′-O-dimethyladenosine(m6Am),2′-O-methyladenosine (Am),N7-methylguanosine (m7G),2′-O-methylguanosine (Gm),N2,N2,7-trimethylguanosine (m2,2,7G),pseudouridine (ψ),N3-methylcytidine (m3C),5-methylcytidine(m5C),2′-O-methyluridine (Um),2′-O-methylcytidine (Cm),and inosine,were clearly detected from mRNA (Fig.1D,Tables S1,S2 and Figs.S2–S4 in Supporting information).The quantification results showed that 6TG treatment led to the significant increase of m1A,Am,Um,Cm,Gm,m7G,m2,2,7G and inosine (Fig.1E and Figs.S5–S7 in Supporting information),suggesting that 6TG could affect the levels of modifications in mRNA.
Since ADAR1 and ADAR2 are responsible for the A-to-I RNA editing,we next further evaluated the expression of ADAR1 and ADAR2 upon 6TG treatment.The result suggested that ADAR2 exhibited significantly increased expression upon 6TG treatment,whereas the expression of ADAR1 had no obvious change (Fig.1F).Thus,we reason that 6TG treatment led to the increased expression of ADAR2,which further caused the increased A-to-I editing in mRNA.
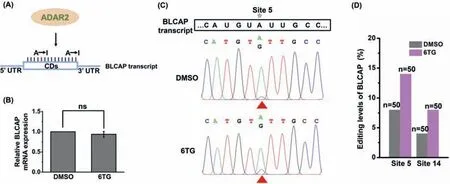
Fig.2.6TG treatment induces the increased A-to-I editing in BLCAP transcript.(A) The schematic illustration showing the A-to-I editing in BLCAP transcript by ADAR2.(B)The relative level of BLCAP in Jurkat-T cells upon 6TG treatment (3 μmol/L for 24 h) compared to control (DMSO treatment).ns,not significant.(C) The A-to-I editing at site 5 in BLCAP transcript with and without 6TG treatment (3 μmol/L for 24 h) by Sanger sequencing.(D) Quantitative evaluation of A-to-I editing at sites 5 and 14 of BLCAP transcript with and without 6TG treatment (3 μmol/L for 24 h) by colony sequencing.
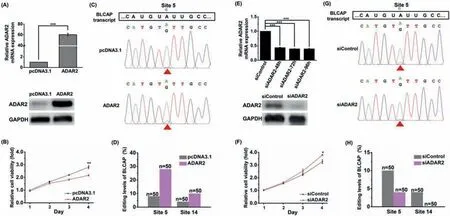
Fig.3.A-to-I editing in BLCAP transcript by ADAR2 modulates cell viability.(A) Evaluation of the overexpression of ADAR2 by qPCR and western blotting.(B) Overexpression of ADAR2 reduced cell viability.(C) The A-to-I editing at site 5 in BLCAP transcript with and without overexpression of ADAR2 by Sanger sequencing.(D) Overexpression of ADAR2 led to the elevated A-to-I editing at sites 5 and 14 of BLCAP transcript by colony sequencing analysis.(E) Evaluation of the siRNA knockdown of ADAR2 by qPCR and western blotting.(F) siRNA knockdown of ADAR2 slightly increased cell viability.(G) The A-to-I editing at site 5 in BLCAP transcript with and without siRNA knockdown of ADAR2 by Sanger sequencing.(H) siRNA knockdown of ADAR2 decreased A-to-I editing at sites 5 and 14 of BLCAP transcript by colony sequencing analysis.*, P<0.05(n=3); **, P<0.01 (n=3); ***, P<0.001 (n=3).
BLCAP is considered as a tumor suppressor gene and it has been reported that BLCAP transcript undergoes multiple A-to-I editing events [45].A general decreased trend in BLCAP-editing level was observed in astrocytomas,bladder cancer and colorectal cancer when compared with normal tissues [46].Some studies revealed that specific substitutions in the coding region can change the functions of the proteins [47].Along this line,the increased level of inosine in mRNA as well as the increased expression of ADAR2 upon 6TG treatment inspired us to further investigate the effect of 6TG on A-to-I editing in the specific BLCAP transcript.
We examined the A-to-I editing in BLCAP transcript with focusing on the sites 5 and 14 of coding region upon 6TG treatment (Fig.2A).Notably,6TG treatment did not alter the expression of BLCAP gene (Fig.2B and Table S3 in Supporting information).Sanger sequencing analysis showed that 6TG treatment led to the increased A-to-I editing at site 5 of BLCAP transcript (Fig.2C).The statistics result from colony sequencing also demonstrated the A-to-I editing at sites 5 and 14 of BLCAP transcript upon 6TG treatment increased from 8% to 14% and 4% to 8%,respectively (Figs.2D and S8 in Supporting information).Collectively,these results revealed 6TG treatment could elevate A-to-I editing at sites 5 and 14 in coding region of BLCAP transcript.
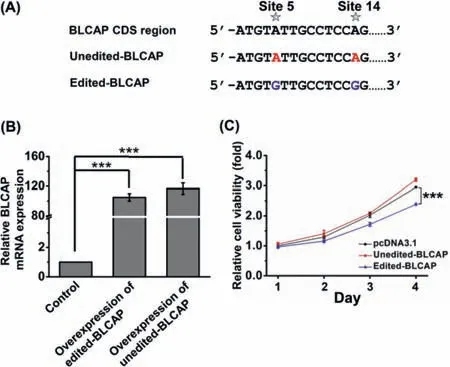
Fig.4.Overexpression of edited-BLCAP reduced cell viability.(A) The schematic illustration of the sequences of two plasmids.The plasmid of unedited-BLCAP contains adenosines at sites 5 and 14.The plasmid of edited-BLCAP contains guanosines at sites 5 and 14.(B) Relative s level upon overexpression of unedited-BLCAP and edited-BLCAP.(C) Effect of overexpression of unedited-BLCAP and edited-BLCAP on cell viability.*** P<0.001 (n=3).
It has been reported that ADAR2 functions as a tumor suppressor gene in brain carcinoma [48] and hepatocellular carcinoma[49].Overexpression of ADAR2 can induce apoptosis of esophageal squamous carcinoma cells [50].Along this line,we hypothesize that increased expression of ADAR2 could elevate A-to-I editing in BLCAP transcript and then eventually induce cell death.In this respect,we first evaluated the cell viability by overexpressing ADAR2 in Jurkat-T cells (Fig.3A).As expected,overexpression of ADAR2 reduced cell viability (Fig.3B).We next analyzed the A-to-I editing of the endogenous BLCAP transcript in ADAR2-overexpressed Jurkat-T cells.Sanger sequencing analysis showed that overexpression of ADAR2 caused the increased A-to-I editing at site 5 of BLCAP transcript (Fig.3C).The statistics result from colony sequencing demonstrated that overexpression of ADAR2 led to the A-to-I editing at sites 5 and 14 increased from 8% to 28% and 4%to 10%,respectively (Fig.3D).Reciprocally,the siRNA knockdown of ADAR2 (Fig.3E) slightly increased cell viability (Fig.3F) and decreased A-to-I editing at sites 5 and 14 of BLCAP transcript (Figs.3G and H).The phenomena of the changed trend of A-to-I editing at sites 5 and 14 of BLCAP transcript through overexpression of ADAR2 was similar to the 6TG treatment.Both the 6TG treatment and overexpression of ADAR2 would increase A-to-I editing at sites 5 and 14 of BLCAP transcript (Figs.2D and 3D) and contribute to cell death.
Given that A-to-I RNA editing substitutes amino acids in BLCAP,we constructed two plasmids for overexpression to further explore the biological significance of BLCAP editing.One plasmid substituted adenosines with guanosines at both sites 5 and 14 (named edited-BLCAP),the other plasmid kept adenosines at both sites 5 and 14 (named unedited-BLCAP) (Fig.4A).The results showed that overexpression of edited-BLCAP (Fig.4B) could reduce cell viability (Fig.4C).However,overexpression of unedited-BLCAP (Fig.4B)did not compromise cell viability (Fig.4C).These results indicated that the A-to-I editing in BLCAP transcript is critical in inducing cell death.However,the mechanism still needs further investigation.
Collectively,our results showed that 6TG treatment could induce the elevated expression of ADAR2,which led to the increased A-to-I editing at sites 5 and 14 of BLCAP transcript and eventually contributed to cell death.This finding indicated a new mechanism for the treatment of acute lymphoblastic leukemia with 6TG.Moreover,the axis of ADAR2-BLCAP could be a potential target for clinical treatment of acute lymphoblastic leukemia in future.Future study on the transcriptome-wide profiling of the A-to-I editing upon 6TG treatment will further promote our understanding of the molecular mechanism of 6TG-induced cytotoxicity through RNA modifications.
In summary,we evaluated the incorporation of 6TG into RNA and investigated its effect on A-to-I editing of BLCAP transcript in acute lymphoblastic leukemia cells.The LC-ESI-MS/MS results showed thatSG could be incorporated into different RNA species,with mRNA harboring the most abundantSG.In addition,we found that 6TG treatment could induce the expression of ADAR2 and therefore elevate the level of inosine modification in mRNA.Specifically,the increased expression of ADAR2 upon 6TG treatment increased A-to-I editing at sites 5 and 14 of BLCAP transcript,which contributed to increased cell death.This study unveils a new mechanism for the treatment of acute lymphoblastic leukemia with using 6TG.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The work is supported by the National Natural Science Foundation of China (Nos.22074110,21635006,21721005).
Supplementary materials
Supplementary material associated with this article can be found,in the online version,at doi:10.1016/j.cclet.2022.01.074.
 Chinese Chemical Letters2023年1期
Chinese Chemical Letters2023年1期
- Chinese Chemical Letters的其它文章
- Diabetic wound healing activated by supramolecular cascade reaction
- MBenes: Two-dimensional transition-metal borides with ordered metal vacancies
- Wet-adhesive materials of oral and maxillofacial region: From design to application
- Diverse catalytic systems for nitrogen-heterocycle formation from O-acyl ketoximes
- Fluorine-containing drugs approved by the FDA in 2021
- The development and application of dual-comb spectroscopy in analytical chemistry
