TaABI19 positively regulates grain development in wheat
LIU Yun-chuan ,WANG Xiao-lu ,HAO Chen-yang ,IRSHAD Ahsan ,LI TianLIU Hong-xiaHOU JianZHANG Xue-yong
1 National Key Facility for Crop Gene Resources and Genetic Improvement,Key Laboratory of Biology and Genetic Improvement of Triticeae Crops,Ministry of Agriculture/Institute of Crop Sciences,Chinese Academy of Agricultural Sciences,Beijing 100081,P.R.China
2 State Key Laboratory of Plant Physiology and Biochemistry,College of Biological Sciences,China Agricultural University,Beijing 100193,P.R.China
3 Institute of Cotton Research,Chinese Academy of Agricultural Sciences,Anyang 455000,P.R.China
4 National Key Facility for Crop Gene Resources and Genetic Improvement/National Center of Space Mutagenesis for Crop Improvement,Institute of Crop Sciences,Chinese Academy of Agricultural Sciences,Beijing 100081,P.R.China
Abstract Starch is the most important component in the endosperm,and its synthesis is regulated by multiple transcription factors(TFs) in cereals.However,whether the functions of these TFs are conserved among cereals remains unclear.In this study,we cloned a B3 family TF in wheat,named TaABI19,based on its orthologous sequence in maize (Zea mays L.).Alignment of the DNA and protein sequences showed that ABI19 is conserved in maize and wheat (Triticum aestivum L.).We found that TaABI19 is highly expressed in young spikes and developing grains,and encodes a nucleus-localized transcriptional activator in wheat.The taabi19-b1 null mutants obtained by EMS exhibited a down-regulation of starch synthesis,shorter grain length and lower thousand-grain weight (TGW).Furthermore,we proved that TaABI19 could bind to the promoters of TaPBF homologous genes and enhance their expression.Haplotype association showed that TaABI19-B1 is significantly associated with TGW.We found that Hap2 and Hap3 were favored and had undergone positive selection in China’s wheat breeding programs.Less than 50% of the modern cultivars convey the favored haplotypes,indicating that TaABI19 still can be considered as a target locus for marker-assisted selection breeding to increase TGW in China.
Keywords: common wheat,transcription factor,haplotype,thousand-grain weight
1.Introduction
Common wheat (Triticum aestivumL.) is the most important staple food in the world,providing a major source of carbohydrates and proteins for the human diet (Brinton and Uauy 2019).Improving the wheat yield is the first objective for most breeders.Thousand-grain weight (TGW) is one of the three major components of yield,and it has high heritability.TGW is determined by grain size and grain filling(Kuchelet al.2007;Fan and Li 2019).
Cereal grain filling is the synthesis of starch,protein,lipid,and other components in the developing endosperm.During grain filling,the glucose is converted to ADPglucose and transported into amyloplasts by the ADPglucose transporter (Brittle1),and then incorporated into starch chains under starch synthase catalysis (Jameset al.2003;Jeonet al.2010;Wanget al.2019).In addition,many transcription factors (TFs),such as OsbZIP58(Wanget al.2013),OsNF-YB1 (Baiet al.2016),OsNFYC12 (Xionget al.2019) etc.in rice (Oryza sativaL.);ZmbZIP91 (Chenet al.2016),ZmNAC128 (Zhanget al.2019) and ZmABI19 (Yanget al.2021) in maize (Zea maysL.);and TabZIP28 (Songet al.2020) and TaNAC019 (Liuet al.2020) in wheat,have been shown to participate in regulating the expression of starch synthesis genes,and thus influence starch synthesis.
TFs ensure precise starch synthesis through protein interaction,coordination or antagonism,and grading in the endosperm.For instance,TFs,such as OsNF-YB1,OsNFYC12 and OsbZIP58,can positively regulate amylose synthesis by activating the expression ofWaxyin rice endosperm (Wanget al.2013;Belloet al.2019;Xionget al.2019).OsNF-YB1 enhances the binding of OsERF115 to the promoters of downstream genes through interaction with OsERF115,which promotes starch synthesis in the endosperm (Xuet al.2016).Some TFs,such as Opaque2 (O2),prolamin-box factor (PBF),OsNAC026 and TaNAC019,have been found to be involved in the synthesis of starch and proteins in developing endosperm,so they work as key factors in maintaining the balance between protein and starch (Schmidtet al.1990;Vicente-Carbajosaet al.1997;Zhanget al.2016;Denget al.2020;Wanget al.2020;Gaoet al.2021).Therefore,cloning and elucidating the functions of TFs in the regulation of starch synthesis and grain development will be beneficial for yield and quality improvements in wheat.Thus far,onlyTabZIP28andTaNAC019have been reported to regulate starch synthesis in wheat (Liuet al.2020;Songet al.2020;Gaoet al.2021).
Gene homology among cereals provides a quick method for obtaining the homologous genes and elucidating their characteristics.The B3 domain-containing TF ZmABI19,a core factor involved in the regulation of starch and protein biosynthesis in endosperm,is necessary for maize grain development and filling (Yanget al.2021).Here,we cloned the orthologous gene ofZmABI19in common wheat and named itTaABI19.We found that TGW and GL were reduced in thetaabi19-b1null mutants.TaABI19 can significantly trans-activate the expression ofTaPBFsthrough binding to their promoters.We found that the haplotypes ofTaABI19were significantly associated with TGW in wheat.
2.Materials and methods
2.1.Plant materials and growth conditions
The Chinese Spring wheat was planted and kept in a germination box at 4°C for one week and then transferred to a plant growth chamber at 24°C with a 16 h light/8 h dark photoperiod.The roots,stems and leaves at different development stages,and grains at 5,10,15,20,25,and 30 days after pollination (DAP),were collected for expression pattern analysis.
TheT.durumwheat mutanttaabi19-b1was planted in the experimental fields of the Institute of Crop Sciences,Beijing,China.Developing grains at 15 DAP were used for RTqPCR analysis,and the developing grains at 20 DAP were used for scanning electron microscopy to observe starch granule development.A natural population containing 157 landraces and 348 modern cultivars,and their phenotype data in multiple environments,were used for haplotype and association analysis (Wanget al.2019).
2.2.RNA isolation and RT-qPCR analysis
Total RNA of collected samples was extracted with RNAiso Plus (TaKaRa,Japan).The first-strand cDNA was synthesized by the Fast Quant RT Kit (Tiangen,China)following the manufacturer’s instructions.The RT-qPCR was performed with TB®Green PremixEx Taq™ (TaKaRa,Japan) on an ABI7300 Real Time PCR System (Applied Biosystems,USA).Each RT-qPCR was repeated three times andTaActinwas employed as the control.The primers are listed in Appendix A.
2.3.Subcellular localization assay of TaABI19
To investigate the subcellular localization of TaABI19,the coding sequences (CDS) ofTaABI19-A1,-B1and-D1were cloned and inserted into the pJIT163-GFP vector.These resultant constructs were transiently expressed in wheat protoplasts,and 35S-GFP was used as the control.The fluorescence signal was measured with a laser confocal microscope (Zeiss LSM 880,Germany) after 16 h incubation under darkness at 24°C.
2.4.Transcriptional activity analysis
To reveal the transcriptional activation of TaABI19,the CDS ofTaABI19-A1/-B1/-D1were inserted into the pGBKT7 vector,and the constructs were then transformed into yeast strain Y2H-Gold.After two days of incubation at 30°C,the yeast was transferred to selective medium without histidine or tryptophan.The pGBKT7 empty vector was used as the control.
2.5.Yeast one hybrid
To reveal the binding ability of TaABI19-B1 to the candidatecis-acting elements and the promoters of the downstream genes,the CDS ofTaABI19-B1was cloned into the pB42AD vector.The DNA fragments of target gene promoters were amplified and cloned into the pLacZi vector.Three repeats of thecis-acting element were inserted into the pLacZi vector,which was transformed into EGY48 strains and cultured on medium containing SD-Trp/Ura/X-gal.The pB42ADTaABI19 and pLacZi empty vector were employed as the negative controls.The primers used for this procedure are listed in Appendix A.
2.6.Dual-luciferase reporter assay
To detect the regulatory effects of TaABI19 on downstream genes,the CDS ofTaABI19-B1was cloned into the effector vector 35S-GAL4DB-Nos,and the promoter sequences of target genes were amplified and fused into the reporter vector pGreenII-0800-LUC (Hellenset al.2005).The effector and reporter vectors were co-transformed intoNicotiana benthamianaprotoplasts at a ratio of 5:1.After incubation in darkness for 16 h at 25°C,the luciferase (LUC)and renilla luciferase (REN) activities were measured by the dual-luciferase reporter assay system (Promega,USA).Each experiment was repeated at least five times.
2.7.KASP marker development and association analysis
For haplotyping of the threeTaABI19homeologous genes in wheat,the target genes in 36 representative collections with high diversity were sequenced.According to the single nucleotide polymorphism (SNP) information,the kompetitive allele specific PCR (KASP) primers were designed to discriminate the variant haplotypes.Subsequently,haplotyping and association analysis was performed in the natural population containing landraces and modern cultivars.
2.8.Statistical analysis
The SPSS 19.0 (IBM,USA) Software was employed to analyze the data.Student’st-test was used to analyze the statistical significance of differences between the wild type (WT) and the mutant in each experimental result.No less than 15 individual plants were used for agronomic trait analysis,and at least three biological repeats were performed for the other experiments.
3.Results
3.1.TaABI19 is mainly expressed in developing grain
TheTaABI19homeologs were located on 3A (Chr3A:467346596–467354585),3B (Chr3B: 448230085–448237201) and 3D (Chr3D: 348429772–348435462) in the Chinese Spring reference genome (IWGSC 2018).TaABI19-A1/-B1/-D1had seven exons and encoded proteins consisting of 301,302 and 301 amino acids,respectively(Fig.1).Multiple sequence alignment analysis suggested that the three TaABI19 homeolog proteins had highly homology to ZmABI19 (Fig.1-B),suggesting that they may be functionally conserved in wheat and maize.

Fig.1 Gene structures and protein sequence alignments of ABI19.A,gene structures of TaABI19 homoeologs and ZmABI19.The black boxes represent exons and polylines represent introns.B,protein sequence alignments of TaABI19 homoeologs and ZmABI19.
The RT-qPCR analysis showed thatTaABI19was constitutively expressed,but predominant in the spike and developing grains (Fig.2-A).A search in the wheat expression profile database showed thatTaABI19-A1,-B1and-D1had similar expression patterns (Appendix B)(Ramirez-Gonzalezet al.2018).The transcriptional activity assay showed that TaABI19 homeolog proteins had transactivation activity (Fig.2-B).The subcellular localization analysis revealed that TaABI19 homeolog proteins were only located in the nucleus (Fig.3).
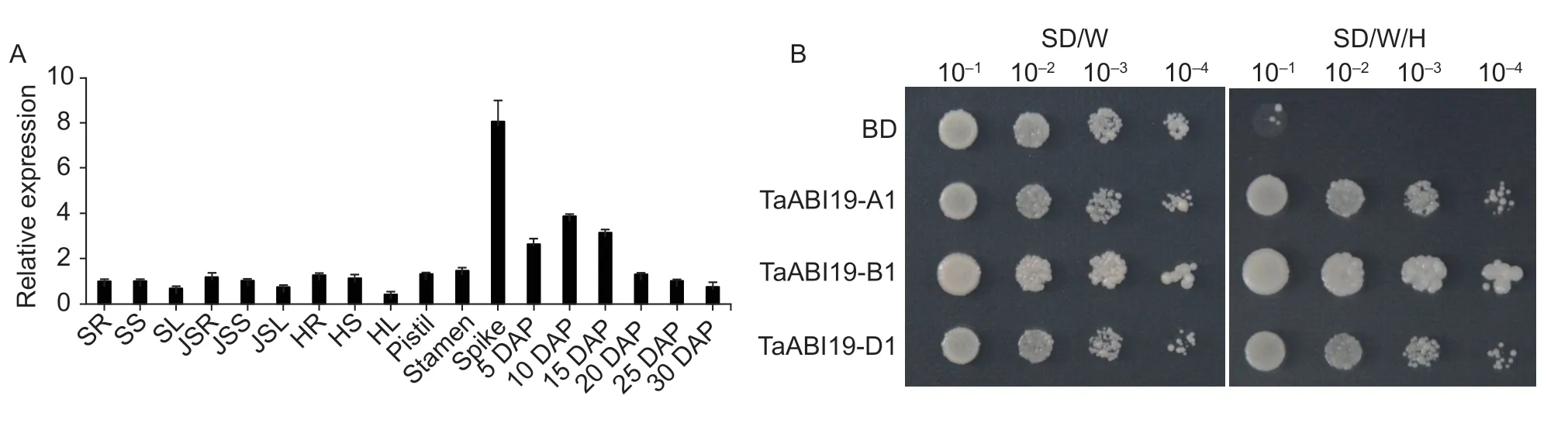
Fig.2 The expression pattern and transcriptional activation of TaABI19. A,the expression pattern of TaABI19.The different tissues of Chinese Spring were used to analyze the expression levels of TaABI19.SR,seedling roots;SS,seedling stems;SL,seedling leaves;JSR,roots at the jointing stage;JSS,stems at the jointing stage;JSL,leaves at the jointing stage;HR,HS and HL represent the roots,stems and leaves at the heading stage,respectively;spike indicates 2 cm young spikes;DAP,day after pollination.TaActin was used as a reference standard.Bars mean SD (n=3).B,the transcriptional activation of TaABI19.The coding sequences of TaABI19 homoeologs were inserted into the pGBKT7 (BD),and then were expressed in the yeast Y2H-Gold strain.These positive strains were tested on the SD/–Trp (SD/W) and SD/–Trp/–His (SD/W/H) media.The empty vector was used as a control.

Fig.3 TaABI19 homologous proteins are localized to nucleus. To analyze the subcellular localization of TaABI19,the TaABI19-A1/-B1/-D1 GFP were transiently expressed in wheat protoplasts.After culturing for 16 h in the dark,the fluorescence signal was observed under a confocal laser microscope.GFP,green fluorescent protein.Bar=10 μm.
3.2.TaABI19 positively regulates grain weight of wheat
To understand the functions ofTaABI19,thetaabi19-b1mutant of durum wheatcv.Kronos was obtained from Dr.Jorge Dubcovsky at UC Davis (California,USA).The mutant site was identified at the “2 377” bp site of ATG downstream forTaABI19-B1;and the “C” to “T” transitions caused a premature translational termination,which produced a protein with no function (Fig.4-A).To detect the further change in expression level caused by the mutation,the grains of 15 DAP were used for RNA extraction and RTqPCR analysis.Compared to the WT,the expression level ofTaABI19was significantly reduced intaabi19-b1,at only about 30% of the WT (Fig.4-D).Notably,the GL oftaabi19-b1was significant reduced (Fig.4-B and F).There was only a minor reduction in grain width (GW) compared with the WT (Fig.4-C and G).Furthermore,the TGW oftaabi19-b1was reduced by 14.7% compared with 45.63 g in WT (Fig.4-E).All these results indicated thatTaABI19-B1positivity regulates GL and TGW in wheat.
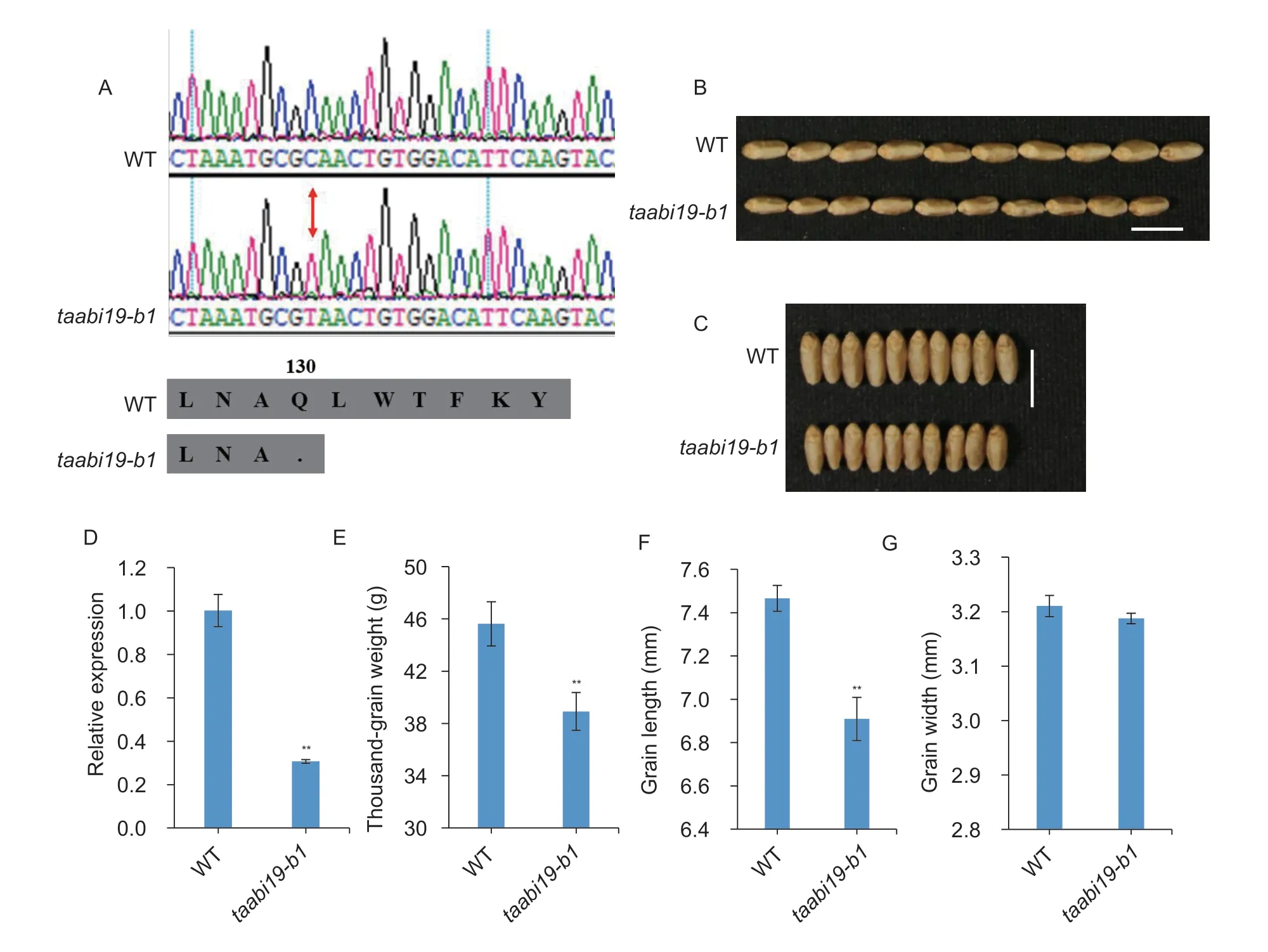
Fig.4 The TaABI19-B1 null mutant has lower grain weight and shorter length. A,sequence identification of the taabi19-b1 null mutant in Kronos wheat.B and C,photographs of grain lengths and widths of taabi19-b1 and wild type (WT).Bar=1 cm.D,expression levels of TaABI19 in WT and taabi19-b1.The data represent the means of three biological replicates.TaActin was used as the reference gene.E–G,means of thousand-grain weight,grain length and width of WT and taabi19-b1.The data represent mean±SD of no less than 15 plants;**,P<0.01 by Student’s t-test.
Electron microscopic scanning showed that the A-type starch granules were slightly smaller intaabi19-b1than the WT in 20 DAP endosperms (Fig.5).This primary implied thatTaABI19is involved in the regulation of starch synthesis in the wheat endosperm.

Fig.5 Scanning election microscopic analysis of endosperm starch. The 20 days after pollination (DAP) grains were used to analyze the phenotypes of starch in wild type (WT) and taabi19-b1.Two biological replicates were performed for each mutant and the WT.Bar=50 μm.
3.3.TaABI19 positively regulates the expression of TaPBF
Previous studies have reported that a RYcis-acting element in the promoter region could be recognized by B3-like TFs (Monkeet al.2004;Penget al.2007).In addition,ZmABI19 was also found to bind the G-box (Yanget al.2021).However,the yeast one-hybrid assay (Y1H) showed that TaABI19 was only binding to the RY motif,but not to the G-box in wheat (Appendix C).Among the threeTaO2homoeologs,onlyTaO2-B1andTaO2-A1had RY motifs in their promoter regions,which could not be recognized and bound by TaABI19 (Appendix D).There was no significant difference in the expression ofTaO2betweentaabi19-b1and the WT (Appendix D).
The RY and G-box motifs were also found in the promoters ofTaPBFs(Fig.6-A),and the core regions were also cloned into a pLacZi vector.After being co-transformed with TaABI19-pB42AD,the co-transformed cells can survive on SD-Trp/Ura/X-gal medium and turn the color to blue(Fig.6-B).This implied TaABI19 could directly bind to the promoter of theTaPBFs.The dual-luciferase reporter system assay (DLR) also supported the idea that TaABI19 can enhance the expression ofTaPBFswith levels nearly two-fold than that of the control (Fig.6-D and E).The RTqPCR results showed that the expression level ofTaPBFintaabi19-b1was significantly lower than that of the WT,further confirming the above results (Fig.6-C).
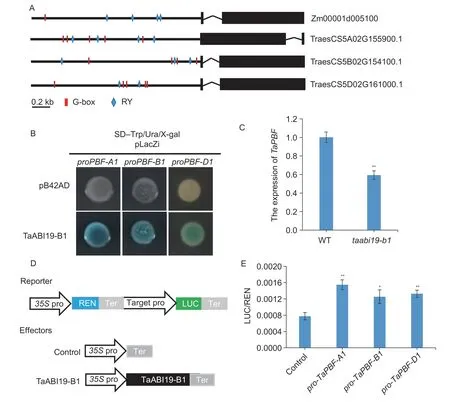
Fig.6 TaABI19 directly regulates the expression of TaPBF. A,gene structures and promoter cis-element analysis of PBF in wheat and maize.B,TaABI19 directly binds to the promoters of TaPBF homoeologs.The TaPBF promoter fragments containing RY and G-box motifs were inserted into the pLacZi vector,and then co-transformed with pB42AD–TaABI19-B1 into the EGY48 yeast strain.SD-Trp/Ura/X-gal was used to detect the binding activity.C,the expression level of TaPBF in 15 days after pollination (DAP) grains of wild type (WT) and taabi19-b1.TaActin was employed as the reference gene.The data represent the means of three biological repeats.**,P<0.01 by Student’s t-test.D,schematic diagrams for the dual-luciferase reporter (DLR) assay.35S pro,CaMV 35S promoter;LUC,firefly luciferase;REN,Renilla luciferase;ter,terminator.E,TaABI19 enhances the activity of TaPBF homoeolog promoters.The reporter and effector plasmids were co-transformed into Nicotiana benthamiana protoplasts,and then the ratios of LUC to REN were determined.The data from five replicates are represented as mean±SD.**,P<0.01 by Student’s t-test.
3.4.Diversity was detected at the TaABI19 homoeolog loci in common wheat
To detect the allelic variations ofTaABI19-A1/-B1/-D1,the coding and 2 kb regions upstream of the start codon in 36 representative cultivars were analyzed (Wanget al.2019).Only one SNP (C/T at the “1 002” bp) was detected in the first intron ofTaABI19-D1,which was not considered in the subsequent association analysis.One SNP and two SNPs were found in the first exon and the promoter region,respectively,which formed two haplotypes atTaABI19-A1(Appendix D).No allelic variation was detected in the CDS ofTaABI19-B1,whereas 21 polymorphic loci,including 19 SNPs and two insertion variant sites,were found in the intron and promoter region ofTaABI19-B1,which formed three basic haplotypes (Fig.7-A).
3.5.TaABI19-B1 is significantly associated with TGW and was strongly selected in Chinese wheat breeding
To analyze the genotype information ofTaABI19-A1in 157 landraces and 348 Chinese modern cultivars,the SNP at the “89” site of the first exon was used to develop the KASP marker and to discriminate the two haplotypes.The phenotypes,including TGW,GL and GW,were used to perform an association analysis of the genotypes.The TGW was significantly different between the two haplotypes ofTaABI19-A1but only in the 2 002 landraces (P=0.005),and the mean of TGW inHap1was 4.65 g higher than inHap2(Appendices E and F).In addition,the GL of the two haplotypes was significantly different in the landraces of different years,but the difference was not significant in the Chinese modern cultivars (Appendix E).However,the difference in GW was not significant between theTaABI19-A1haplotypes (Appendix E).
Two KASP markers were employed to distinguish the three haplotypes ofTaABI19-B1.Association analysis was preformed among the genotypes and phenotypes,including TGW,GL and GW.The TGW was significantly associated with theTaABI19-B1haplotypes.Among landraces,the mean TGW ofHap2was significantly higher than that ofHap1in all environments by 10.75% in 2002 Luoyang (LY),Henan Province,16.9% in 2005 LY,14.05% in 2006 Xinxiang(XX),Henan Province and 9.95% in 2010 Shunyi (SY),Beijing (Fig.7-B;Appendix G).Similarly,the TGW ofHap2was also significantly higher than that ofHap1in the modern cultivars by 4.9% in 2002 LY (P=0.044),5.53% in 2005 LY (P=0.028) and 4.86% in 2010 SY (P=0.05) (Fig.7-C;Appendix G).The mean TGW ofHap3was notably higher than that ofHap1only in the landraces by 13.38% in 2002 LY (P=0.005),8.39% in 2006 XX (P=0.048) and 11.38% in 2010 SY (P=0.014) (Fig.7-B;Appendix G).Besides,the GL in landraces was also significantly associated with theTaABI19-B1haplotypes.The mean GL values ofHap2andHap3were higher than that ofHap1(Appendix H).In the landraces,the average GL ofHap2was significantly longer than that ofHap1by 5.34% 2002 in LY,5.44% in 2006 XX and 4.22% in 2010 LY (Appendix G).In addition,the mean GL ofHap3was also notably longer than that ofHap1(P<0.01) (Appendix G).Furthermore,the mean GW ofHap2in landraces was significantly wider than that ofHap1in three environments (P<0.05) by 6.1% in 2002 LY,4.13% in 2005 LY and 4.08% in 2006 XX (Appendices G and H).The above results were consistent with the phenotypes of thetaabi19-b1mutant,which indicate thatTaABI19positively regulates TGW and GL.
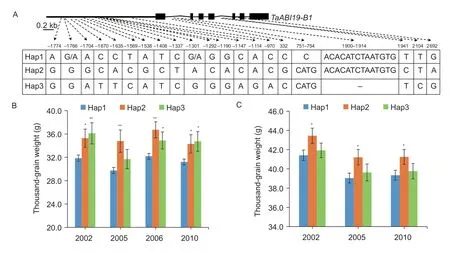
Fig.7 TaABI19-B1 haplotypes are associated with thousand-grain weight. A,haplotypes of TaABI19-B1.The polymorphic sites and variation information are indicated.B,mean thousand-grain weight of TaABI19-B1 haplotypes in the landraces.C,mean thousand grain weights of TaABI19-B1 haplotypes in the modern cultivars.For B and C,the x-axis represents different environments (Luoyang,2002;Luoyang,2005;Xinxiang,2006;Shunyi,2010).Bars mean SD.Significance analysis was carried outbyStudent’st-test,**,P<0.01;*,P<0.05.
The TGW values ofTaABI19-B1-Hap2and-Hap3were significantly higher than that ofTaABI19-B1-Hap1,so we further analyzed whether they were under selection during Chinese wheat breeding.The results showed thatTaABI19-B1-Hap2and-Hap3had undergone positive selection in several ecological wheat zones,especially in the major production zones I,II,III and VII (Fig.8).
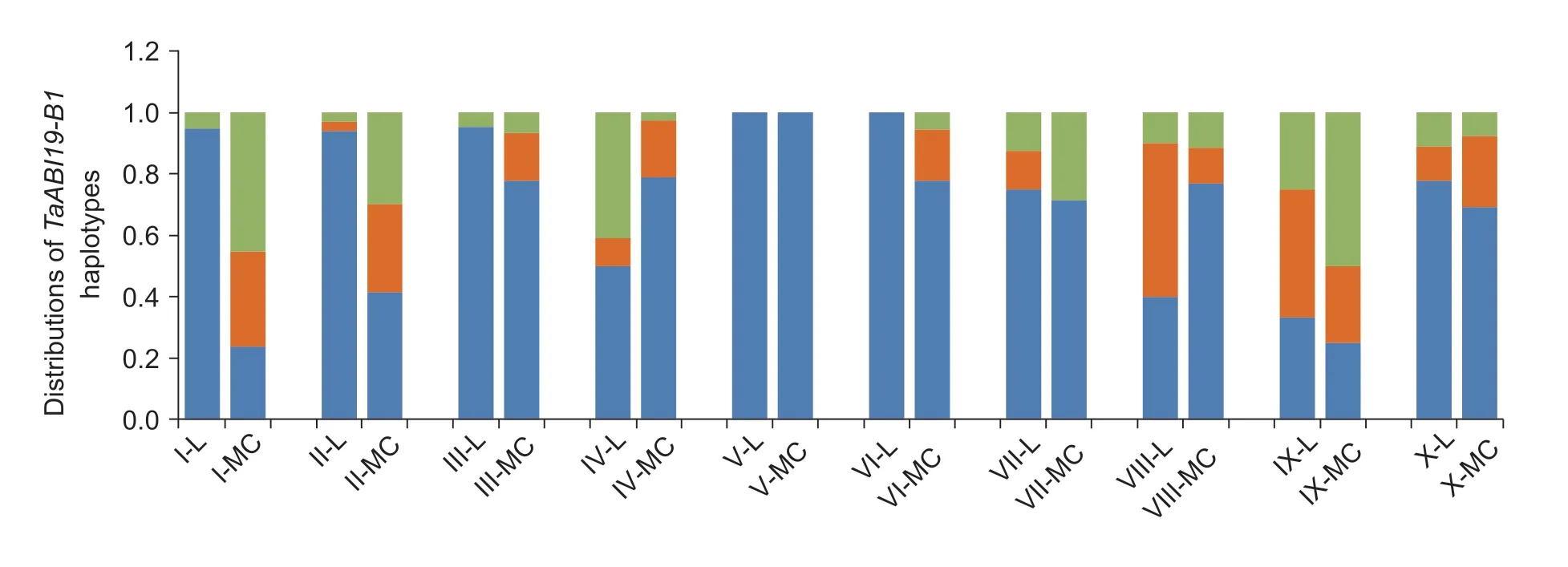
Fig.8 Distributions of TaABI19-B1 haplotypes in 10 wheat ecological regions in China.L,landraces;MC,modern cultivars.I,northern winter wheat region;II,Yellow and Huai River valley winter wheat region;III,low and middle Yangtze River valley winter wheat region;IV,southwestern winter wheat region;V,southern winter wheat region;VI,northeastern spring wheat region;VII,northern spring wheat region;VIII,northwestern spring wheat region;IX,Qinghai–Tibet spring–winter wheat region;X,Xinjiang winter–spring wheat region.
4.Discussion
4.1.TaABI19 is a positive regulator of wheat grain length and grain weight
Grain development and filling are important for cereal yields(Olsenet al.1991;Daiet al.2021).The loss of functions of these genes regulating grain development and filling lead to defective grain development,resulting in a significantly reduced yield (Fenget al.2018;Daiet al.2021).Multiple B3 TFs have been shown to be involved in the regulation of seed development and the synthesis of seed storage substances(Suzuki and McCarty 2008).VP1is indispensable for scutellum development and protein reallocation,and a loss of function leads to a misshapen scutellum in maize (Zhenget al.2019).ZmABI19can regulate grain development and filling by enhancing the expression ofVP1,O2,PBF,and others,and theZmABI19null mutant showed abnormal endosperm and reduced starch synthesis (Yanget al.2021).Here,we found thatTaABI19was mainly expressed in developing wheat grains (Fig.2-A).Thetaabi19-b1null mutant showed lower values of TGW and GL (Fig.4).Scanning electron microscopy showed that the synthesis of starch was slightly impeded and the starch granules were smaller in thetaabi19-b1mutant (Fig.5).However,taabi19-b1did not produce a severe grain developmental defect phenotype similar to that ofzmabi19.Due to the polyploid characteristics of common wheat,the dose or compensation effect ofTaABI19homologues may lead to the differences in the mutant phenotypes between maize and wheat (Chiaet al.2020;Hawkinset al.2021).In the future,the functions ofTaABI19in regulating grain development and filling will be analyzed more comprehensively and accurately by using gene knockout or a homozygous genetic background oftaabi19-b1.
4.2.Variations within the promoters of the downstream gene led to ABl19 regulatory differentiation between maize and wheat
As upstream regulatory genes,TFs regulate grain development and filling (including starch and protein synthesis,etc.),and ultimately affect grain yield.However,the research in wheat lags behind the efforts in rice and maize.Verifying whether the function of TFs is fully conserved or partially differentiated among cereals will help to quickly clarify the regulation of wheat grain development and filling.In maize,ZmABI19 can directly bind to the G-box and RY motif in the promoter regions ofO2andPBFand trans-activate their expression (Yanget al.2021).Here,we found that TaABI19 only bound to the RY motif (Appendix C),and the RY motifs were found in the proximal promoters of threeTaPBFhomoeologs(Fig.6-A).Furthermore,the Y1H,DLR and RT-qPCR results showed that TaABI19 could only bind to the promoters ofTaPBFand activate its expression (Fig.6-B and C),which was consistent with the effect of ZmABI19(Yanget al.2021).Although the RY element exists in the proximal region of theTaO2-B1promoter and the distal region of theTaO2-A1promoter,TaABI19 did not bind to and enhance its expression (Appendix D),which is different from the regulatory mechanism of ZmABI19 (Yanget al.2021).As central regulators in maize grain filling,O2 and PBF can enhance the expression of multiple genes involved in endosperm development,sucrose cleavage and starch synthesis in endosperm,thus affecting grain yield and quality (Schmidtet al.1990;Vicente-Carbajosaet al.1997;Zhanget al.2016;Zhanet al.2018;Denget al.2020).ZmABI19 is an upstream TF ofO2andPBF,and the loss of function of ZmABI19 resulted in severe impairment of maize grain developmental filling.In this work,thetaabi19-b1null mutant did not lead to stunting phenotypes as notable as those in maize,suggesting thatABI19is partially functionally differentiated between maize and wheat,and this differentiation might be caused by the variations ofcis-elements in the core region of the downstream gene promoter.Combined with the results of this work,we speculated that the functional differentiation of orthologous TFs among cereals might be caused by the variations ofcis-elements in the promoter region.In the future,exploring the variations in the promoter and 5´-UTR regions of starch synthesis and related TFs genes in cereal crops will be helpful for better understanding endosperm development and grain filling.
4.3.TaABI19-B1 will be a new selection site in wheat breeding
In this study,three haplotypes were detected atTaABI19-B1,among whichHap2andHap3are the favored haplotypes for breeding with higher TGW and GL (Fig.7).Grain weight,as the one of the three factors of yield,is an important selection target for wheat breeders and has contributed the most to wheat yield improvements in the past.In previous studies,the grain weight genes have been extensively studied,such asGW2(Suet al.2011;Maphosaet al.2014),DA1(Liuet al.2019),GW7(Wang Wet al.2019) andSus2(Houet al.2014),and their superior haplotypes have been positively selected in Chinese wheat breeding.Here,we found thatTaABI19-B1-Hap2and-Hap3with higher TGW were significantly selected,especially in the I,II,III and VII wheat production regions in China (Fig.8).However,the proportions of other haplotypes were still very high in regions V,IV and VIII (Fig.8).This means that the favored haplotypes have not been fully positively selected in each wheat region.The KASP markers that were developed based on the haplotypes will likely be used in future wheat marker-assisted breeding.
5.Conclusion
This study shows thatTaABI19is a key factor in the regulation of grain weight and filling.Thetaabi19-b1null mutant has lower grain weight and grain length.TaABI19can promote the expression ofTaPBF,while it has no effect on the expression ofTaO2.The favored haplotypes ofTaABI19have higher TGW,and are likely to serve as a new selection locus for wheat yield breeding.
Acknowledgements
The Kronos EMS mutant lines were developed by Dr.Jorge Dubcovsky at University of California,Davis (California,USA).The authors are very grateful to Prof.Wu Jiajie(Shandong Agricultural University,State key Laboratory of Crop Biology) for providing thetaabi19-b1mutant wheat.This work was supported by the the Central Public-interest Scientific Institution Basal Research Fund,Chinese Academy of Agricultural Sciences (Y2017PT39).
Declaration of competing interest
The authors declare that they have no conflict of interest.
Appendicesassociated with this paper are available on http://www.ChinaAgriSci.com/V2/En/appendix.htm
 Journal of Integrative Agriculture2023年1期
Journal of Integrative Agriculture2023年1期
- Journal of Integrative Agriculture的其它文章
- Advances in studies on the physiological and molecular regulation of barley tillering
- Antioxidant lignans sesamin and sesamolin in sesame (Sesamum indicum L.): A comprehensive review and future prospects
- Genetic effects of Agropyron cristatum 2P chromosome translocation fragments in a wheat background
- Grain yield,nitrogen use efficiency and physiological performance of indica/japonica hybrid rice in response to various nitrogen rates
- Border effects of the main and ratoon crops in the rice ratooning system
- Agronomic management practices in dryland wheat result in variations in precipitation use efficiency due to their differential impacts on the steps in the precipitation use process
