Evaluation of the metabolism of PEP06,an endostatin-RGDRGD 30-amino-acid polypeptide and a promising novel drug for targeting tumor cells
Liyun Niu,Huiyu Zhou,b,Yueru Lin,Y Go,Yulu Liu,Ruoln Gu,Zhuon Wu,Xioxi Zhu,Hui Gn,Zhiyun Meng,**,Guifng Dou,*
aState Key Laboratory of Drug Metabolism and Pharmacokinetics,Department of Pharmaceutical Sciences,Beijing Institute of Radiation Medicine,Beijing,100850,China
bHenan University,Kaifeng,Henan,475004,China
ABSTRACT
PEP06 is a novel endostatin-Arg-Gly-Asp-Arg-Gly-Asp(RGDRGD)30-amino-acid polypeptide featuring a terminally fused RGDRGD hexapeptide at the N terminus.The active endostatin fragment of PEP06 directly targets tumor cells and exerts an antitumoral effect.However,little is known about the kinetics and degradation products of PEP06 in vitro or in vivo.In this study,we investigated the in vitro metabolic stability of PEP06 after it was incubated with living cells obtained from animals of different species;we further identified the degradation characteristics of its cleavage products.PEP06 underwent rapid enzymatic degradation in multiple types of living cells,and the liver,kidney,and blood play important roles in the metabolism and clearance of the peptides resulting from the molecular degradation of PEP06.We identified metabolites of PEP06 using full-scan mass spectrometry(MS)and tandem MS(MS2),wherein 43 metabolites were characterized and identified as the degradation metabolites from the parent peptide,formed by successive losses of amino acids.The metabolites were C and N terminal truncated products of PEP06.The structures of 11 metabolites(M6,M7,M16,M17,M21,M25,M33,M34,M39,M40,and M42)were further confirmed by comparing the retention times of similar full MS spectrum and MS2spectrum information with reference standards for the synthesized metabolites.We have demonstrated the metabolic stability of PEP06 in vitro and identified a series of potentially bioactive downstream metabolites of PEP06,which can support further drug research.
Keywords:
PEP06
HRMS
Metabolic stability
Metabolites identification
Peer review under responsibility of Xi'an Jiaotong University.
1.Introduction
Cancer is widely recognized as a major global public health problem,and it is,unfortunately,a leading cause of death worldwide[1-3].Despite the increasing incidence of cancer and cancerrelated deaths,significant progress has been made in cancer screening methods and in surgery,chemotherapy,radiotherapy,and immunotherapy used to treat cancer[4].
Peptides and other biologics have gained increasing attention as potential antitumor drugs during the past couple of decades due to their unique pharmacological properties and intrinsic characteristics[5-7].Small peptides are ideal candidates for novel drug development owing to their low cost of production and reduced complexity compared with typical protein-based biopharmaceuticals,such as monoclonal antibodies or cytokines.Further,they typically cost as much as small organic molecules.Given these features,peptides are truly in the sweet spot when it comes to the choice between biopharmaceuticals and small molecules for drug development.The development of novel and more effective peptides is subsequently attracting much interest in the research community.
Angiogenesis,which involves the recruitment of new capillary blood vessels,is thought to play a major role in the growth and development of human tumors,as this process promotes nutrient-limited tumor growth and metastasis[8].Numerous studies have shown that angiogenesis is a complex process regulated by multiple proteins,including integrins.Integrins are heterodimeric transmembrane glycoproteins that execute significant regulatory functions in both normal and tumor cells[9].Intensive investigation of the functions of several integrins has led to the development of robust novel antitumor antagonists of integrins.Integrin αvβ3[10,11],an angiogenic vascular tissue marker,was found overexpressed in tumor-associated blood vessels,such as activated endothelial cells,but not in resting vessels.Therefore,drugs,such as peptides and antibody antagonists,that block integrin αvβ3may effectively block tumor angiogenesis and tumor growth.
Arg-Gly-Asp(RGD)has been recognized as an essential peptide binding motif for αvβ3receptor[12].Since integrins can specifically recognize the RGD sequence,the high affinity of RGD peptides for αvβ3receptor facilitates their potential use in cancer diagnosis and therapy[13].Hence,a promising strategy for RGD-modified drug development focuses on targeting tumorassociated blood vessels.Researchers have made significant progress in developing RGD-modified peptides that target tumor blood vessels[14].
Endostatin,a 183-amino-acid peptide cleavage fragment corresponding to the C terminus of gelatin 18,possesses a significant antitumor activity as observed from its inhibitory effect on tumor angiogenesis[15-18].Further research revealed that the 27-amino-acid peptide derived from N terminus of endostatin has a comparable antitumor activity with that of full-length endostatin.We focused on a similar but novel 30-amino-acid polypeptide endostatin-RGDRGD,i.e.,PEP06[19].PEP06 is synthesized by terminally fusing the RGDRGD hexapeptide at the N terminus with the active fragment of endostatin.This modification of PEP06 peptide comprising a motif of two RGD sequences attached to the N terminus supports the direct targeting of tumor angiogenesis,with high antimetastatic activity and a high production yield.The NH2group,attached to the C terminus of the PEP06 peptide,is artificially amidated to stabilize the synthetic peptide structure.
The potent antitumor effect of PEP06 both in vitro and in vivo has already been demonstrated[20].In addition,PEP06 is proven to be effective against colorectal cancer.Preclinical pharmacokinetic studies[21]of PEP06 show that it has a short elimination half-life of 0.05 h in rats at a dose of 30 mg/kg and 0.13 h in cynomolgus monkeys at a dose of 15 mg/kg.PEP06 has progressed to phase I clinical trials in China.
To better understand its pharmacologic properties,we focused on evaluating the metabolism and degradation of PEP06 within different living cells obtained from animals of different species in vitro and characterizing the metabolites using liquid chromatography-tandem mass spectrometry(LC-MS/MS).Our in vitro findings show that the PEP06 metabolites may be explained,at least in part,by their short half-life during the stability study.Moreover,we describe the development and optimization of a reliable method using ultra-performance liquid chromatography coupled to high-resolution mass spectrometry(UPLC-HRMS)for the detection of these peptide metabolites,thus facilitating the determination of the specific metabolic cleavage sites and specific metabolites generated.
We subsequently identified and characterized the PEP06 metabolites from different cell types of several species.To the best of our knowledge,this is the first report on the identification and characterization of specific PEP06 metabolites from in vitro metabolic experiments.Finally,the peptides from the more abundant metabolites were synthetized to confirm their proposed structures.
2.Experimental
2.1.Chemicals and reagents
Standard preparations of PEP06(Fig.1)and its IS PEP10(RGDRGDMHSHRDFQPVLHLVALNSPLSGGMGG)were synthesized by Chengdu Shengnuo BioTec Co.,Ltd.(Chengdu,China).LC-MS-grade methanol,acetonitrile,and isopropanol were purchased from Thermo Fisher Scientific Inc.(Shanghai,China).In addition,LC-MS-grade formic acid(FA)and acetic acid(AA)were obtained from Sigma-Aldrich(St.Louis,MO,USA).Ultrapure water was purified using a Milli-Q system(Millipore,Milford,MA,USA).
2.2.Experimental animals
Sprague-Dawley(SD)rats were purchased from Beijing KeYu Animal Breeding Center(Beijing,China),and the license for the production of experimental rats is SCXK(Jing)2018-0010.Cynomolgus monkeys were purchased from Beijing Prima Biotech Inc.(Beijing,China),and the license for the production of experimental monkeys is SCXK(Jing)2018-0001.Bama miniature pigs were purchased from Beijing ShiChuangShiJi Minipig Breeding Base(Beijing,China),and the license for the production of experimental pigs is SCXK(Jing)2018-0011.All the animal experiments were approved by the Animal Care and Welfare Committee of the Institute of Radiation Medicine,the Academy of Military Medical Sciences(Beijing,China),and were in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
2.3.Preparation of biological matrices
2.3.1.Preparation of fresh whole blood
Blood samples from SD rats,cynomolgus monkeys,Bama miniature pigs,and humans were collected and transferred to heparinized tubes,and the blood samples were inverted 3-4 times for homogeneous mixing.The prepared blood matrices were stored in tubes at-80°C until they were needed for the experiments.
2.3.2.Preparation of liver and kidney homogenates
SD rats,cynomolgus monkeys,and Bama miniature pigs were sacrificed by an overdose of sodium pentobarbital,and tissue materials were immediately excised from these animals.Tissues were placed in ice-cold 0.9% saline and washed to remove the remaining blood.The chilled tissues were weighed,chopped,and homogenized at a ratio of 1:4(gram of tissue/mL of 0.9% saline).The homogenates were centrifuged at 4°C and 1,000 g for 10 min.Then supernatants were collected and stored at-80°C until needed for experimental analysis.
2.4.Metabolic stability of PEP06 in vitro
A stock solution of PEP06 was prepared in 2% AA at 1 mg/mL and diluted to 20μg/mL in individually prepared multiple biological matrices.Fresh blood,liver,and kidney homogenates from different species were incubated in a water bath set to 37°C with continuous shaking when PEP06 was added.Each sample was carried out in triplicate sets.Samples were collected regularly at 0 min and at frequent intervals of 1,3,5,10,15,20,30,45,60,120,and 180 min during incubation.Aliquots(50μL)of the reaction mixture were added into a 1.5 mL Eppendorf tube,and peptides were precipitated in methanol:acetonitrile(1:1,V/V)containing 0.3% FA,which were used at volumes six-fold those of aliquots samples.PEP10 was serially diluted to final concentration of 25μg/mL with methanol,and 5μL was added to the mentioned samples as the internal standard.The whole process was performed in an ice bath for a maximum of 30 min.The samples were vortexed to precipitate the protein and then centrifuged at 13,201 g for 15 min at 4°C.Supernatants were carefully transferred to a new tube and doublediluted with water containing 0.3% FA.Finally,the mentioned samples above were centrifuged at 13,201 g for 15 min at 4°C,and the supernatants after secondary precipitation were injected onto the liquid chromatography-mass spectrometry for quantification.

Fig.1.The amino acid sequence of PEP06.
2.5.Metabolic identification of PEP06 in vitro
Fresh whole blood,liver,and kidney homogenates were temperature-equilibrated at 37°C in a water bath for 5 min with shaking.Then,the reaction mixture of PEP06 peptide was incubated at a final increased concentration of 1 mg/mL for the purpose of performing qualitative metabolite research.For whole blood,a biological aliquot was withdrawn after 15 min of incubation from SD rats,and cynomolgus monkeys;for liver or kidney homogenates,aliquots were withdrawn after 15 min of incubation from SD rats,cynomolgus monkeys,and Bama miniature pigs or aliquot was drawn from blood after 30 min of incubation from Bama miniature pigs and humans.The reactions were conducted for a suitable duration based on the metabolic stability of PEP06 in vitro,which was longer than at least two half-lives individually.The samples were precipitated using 300μL of methanol:acetonitrile(1:1,V/V)containing 0.3% FA immediately and vortexed and centrifuged at 13,201 g for 15 min at 4°C;the supernatant was then diluted by adding an equal volume of water containing 0.3% FA.The diluted samples were vortexed for 2 min and centrifuged at 13,201 g for 15 min at 4°C;the final supernatants were then added to autosampler vials to identify the PEP06 metabolites by ultra-performance liquid chromatography-mass spectrometry(UPLC-MS).
2.6.Chromatographic and mass spectrometry conditions
2.6.1.LC-MS/MS methodology for quantification of PEP06
All biological samples for evaluating the metabolic stability of PEP06 were analyzed by high-performance liquid chromatographymass spectrometry(HPLC-MS),performed on an API4000 triple quadrupole mass spectrometer(Applied Biosystem,Framingham,MA,USA)equipped with a Nanospace SI-2 chromatography system(Shiseido,Tokyo,Japan).The detailed quantitative methods used have been previously reported[21].The dynamic range of analysis with the structural analogues as the internal standard was validated between 10 and 2,000 ng/mL.Data were acquired and processed by Analyst 1.6.3(Applied Biosystem,Framingham,MA,USA).
2.6.2.LC-MS/MS methodology for metabolite identification studies
PEP06 and its metabolites were analyzed on a UPLC-MS/MS system consisting of a Dionex UltiMate 3000,Ultimate 3000 Autosampler equipped with a Q-Exactive Orbitrap™mass spectrometer,all of which were purchased from Thermo Fisher Scientific Inc.(Waltham,MA,USA).Chromatography separation was achieved on a Kinetex UPLC peptide BEH C18column(2.1 mm× 100 mm,2.6μm,100 Å;Phenomenex,Torrance,CA,USA)using 0.3%(V/V)aqueous formic acid and acetonitrile containing 0.3%(V/V)formic acid as mobile phase A and B,respectively,lf owing at 300 μL/min;the injection volume was 5 μL.A 50-min binary gradient elution was used to separate metabolites completely.The multi-step gradient started with 2% B for 5 min and increased linearly to 30% B over 35 min and then in the following 1 min increased to 80% B quickly.The mobile phase gradient was held at 80% B during 36-45 min,reduced to 2% B in 1 min,and held for 4 min,with a total duration of 50 min.MS analyses were performed on a Q-Exactive Orbitrap™high-resolution mass spectrometer equipped with an electrospray ionization(ESI)probe.The Q-Exactive MS was tuned and calibrated once a week in both positive and negative modes.In the process of qualification of PEP06 and its metabolites,a full scan/data-dependent MS/MS(FS-dd-MS/MS)scan mode was adopted.Full-scan MS data was acquired at a mass scan range of 200-2,000 m/z,setting a resolution of 70,000 and chromatogram peak width of 6 s.The dd-MS2was performed at a resolution of 35,000 with an isolation width of 1.8 m/z.Furthermore,stepped normalized collision energy(NCE),consisting of three different NCE values:20,40,and 60,was used for the same MS/MS event to create more fragments.The sheath gas flow rate and auxiliary gas flow rate were set at 45 arbitrary units and 10 arbitrary units,respectively.The spray voltage,temperature of the capillary,S-lens RF level,and Aux gas heater temperature were set as 3.5 kV,350°C,55 arbitrary units and 400°C,respectively.Data acquisition was facilitated by Xcalibur software version 4.0.27.13(Thermo Fisher Scientific Inc.,Waltham,MA,USA).
2.7.Verification of the identity of the metabolites
In this study,11 standard metabolites were synthesized for verification of the generated metabolites.The amino acid sequences of the generated metabolites were verified by comparing the retention time,full scan MS spectra,and MS/MS fragment ions with those of the synthesized standard metabolites.The 11 standard metabolites were diluted to 50μg/mL and injected to the QExactive MS.
2.8.Data analysis
The substrate remaining percentage(%)was calculated by comparing the peak area-ratio of the detected compound with that of the IS at each time point to time zero.The elimination rate constant(ke)of PEP06 in in vitro metabolic studies involving different biological matrices was determined by the natural logarithm regression analysis of remaining percentage(%)versus time points.Then,substrate disappearance(t1/2)in vitro was obtained using the following equation:t1/2=0.693/ke.
The resulting peptide mass spectrum was compared with that in the databases of ProteinProspector Tools(University of California,San Francisco,CA,USA)to elucidate the amino acid sequences of generated metabolites.
3.Results and discussion
3.1.Metabolic stability of PEP06 in vitro
PEP06 metabolic stability studies in biological matrix were conducted in blood,liver homogenates,and kidney homogenates obtained from different species.The liver and kidney both had an extremely important effect on the metabolism and clearance of molecular peptides.In addition,blood was also confirmed to play a significant role in the metabolism of peptides.PEP06 was degraded in vitro in blood obtained from cynomolgus monkeys,Bama miniature pigs,and humans with t1/2of(0.604±0.008)min,(14.875±0.498)min,and(13.909±0.525)min,respectively(Table1).PEP06 rapidly metabolized in rats within 1 min of incubation;theremaining percentage(%)was less than 5%,and therefore,the half-life could not be calculated currently.The degradation rate of blood from different species was as follows,in an ascending order:Bama miniature pigs<humans<cynomolgus monkeys<SD rats(Fig.2).The t1/2of PEP06 in liver homogenates of SD rats,cynomolgus monkeys, and Bama miniature pigs was(2.069±0.022)min,(3.229±0.100)min,and(3.775±0.134)min,respectively.The degradation rate was SD rats>cynomolgus monkeys>Bama miniature pigs.Finally,the degradation rate of PEP06 in kidney homogenates of SD rats,cynomolgus monkeys,and Bama miniature pigs was determined considering t1/2of(1.833±0.068)min,(1.298±0.035)min,and(1.715±0.143)min,respectively.The degradation rate in kidney homogenates was cynomolgus monkeys>Bama miniature pigs>SD rats.
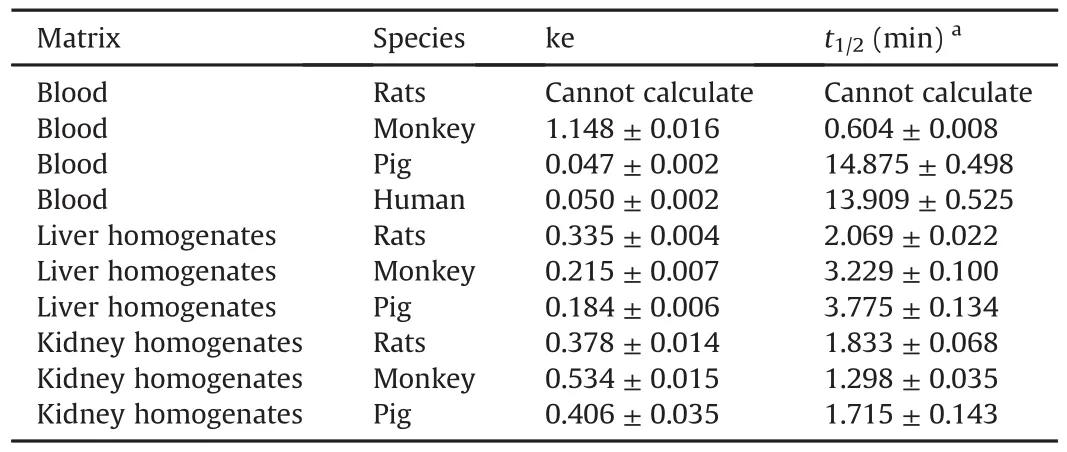
Table 1In vitro stability of PEP06 in blood,liver,and kidney from different species.
3.2.Development of a method to identify PEP06 metabolites in vitro
UPLC-MS is a vital method for the quantitative and qualitative analyses of metabolism and pharmacokinetics for many contemporary drugs,including small molecules,peptides,and others.However,the identification of these metabolites poses a comparably new challenge for peptides compared to routine small molecules.
3.2.1.Optimization of liquid chromatography
UPLC,in particular,gives higher chromatographic resolution,results in sharper peaks,and is more sensitive than HPLC.A common strategy for optimizing an LC method involves sample preparation,selection of chromatographic columns,and gradient distribution of the mobile phase.In this study,the samples prepared in ice bath were used for quantifying and identifying metabolites.This sample preparation process preserved sample stability and reduced the matrix effect.Reverse-phase LC support for the separation of peptides was thought ideally suited for peptide chromatographic analysis in most cases.To develop a UPLC method,columns were chosen to obtain high responses and acceptable peak shapes.Two columns,Biobasic C18column(2.1 mm×100 mm,5μm;Thermo Fisher Scientific Inc.,Waltham,MA,USA)and Kinetex C18LC column(2.1 mm×100 mm,2.6μm,100 Å;Phenomenex,Torrance,CA,USA),were screened for response,peak shape,and interference.The Kinetex C18LC column showed higher response,better peak shapes,and better resolution.Like in many other peptides,PEP06-derived metabolites were present in trace concentrations,thus needing gradient optimization,which was performed in a series of multi-step elution adjustments.Finally,a slow gradient method was developed.The multi-step gradient of method was initiated with 2% organic phase for 5 min and then the proportion of organic phase increased at a slow slope to 30% within 5-35 min.At this stage,most metabolites were eluted.The proportion of organic phase was increased to 80% in 1 min to elute matrix interference or others and maintained for several minutes to ensure parallelism between each sample injection.
3.2.2.Optimization of mass spectrometry
Mass spectrometry-based approaches play a substantial role in the analysis,quantification,characterization,and identification of metabolites,considering the developments in peptide and protein drug studies over the past 40 years[22-26].For peptides and protein drugs,ESI is the most widely used ionizer as it is an LC-compatible ionization/interface method coupled mass spectrometers to chromatographs operated under essentially standard LC conditions for flow and separation.In this study,samples were acquired by Q-Exactive MS,which was the first MS instrument that combined quadrupoles as the parent ion selectivity with high resolution orbitrap mass spectrometry.Although time of flight(TOF)MS has certain advantages in terms of scanning speed,Q-Exactive Orbitrap™MS has near-perfect performance in other aspects,such as mass axis stability,resolution,and reproducibility.All the precursor ions should ideally fragment and give a product ion spectra beneficial for identifying metabolites.However,it is challenging to ensure sufficient precursor ions to produce a sufficiently large number of ions with only one single collision energy(CE).“One size fits all”is not suitable for all peptides due to the diversity in peptide sequence.To acquire a large number of useable ions,and therefore facilitate the structural elucidation of metabolites,stepped NCE was used for MS2at low,medium,and high collisional energies,and fragments from different fragmentation events were combined and detected simultaneously without sacrificing the throughput.Therefore,the multiple collision energy mode provided more identifiable product ions,improved fragmentation efficiency,and yielded a greater number of low intensity and identifiable fragments compared with a single CE.
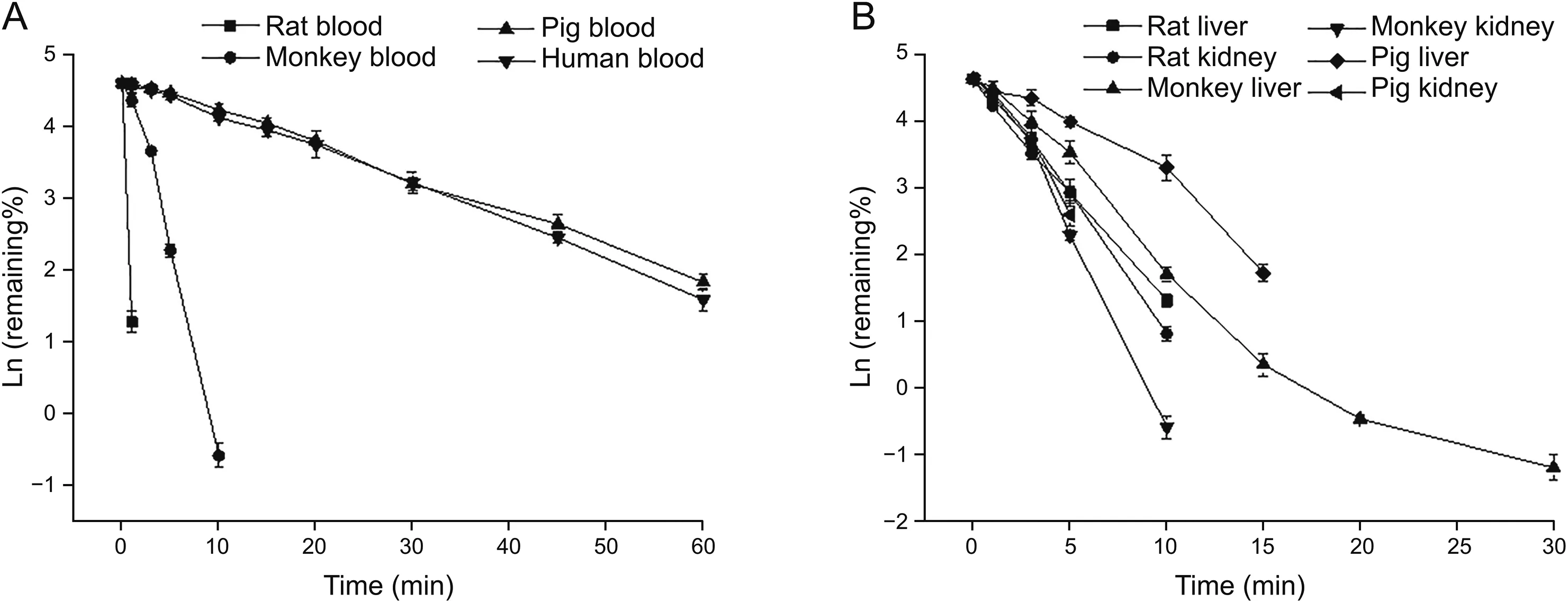
Fig.2.Time course for the in vitro degradation of PEP06 upon incubation at 37°C.(A)PEP06 degradation in blood obtained from animals of different species.(B)PEP06 degradation in the liver and kidney homogenates obtained from animals of different species.
3.3.Metabolic identification of PEP06 in vitro
HRMS has become an increasingly important tool for investigating peptides or proteins.The peptide molecular weight was determined by interpreting the mass spectra of primary structure.For ion fragmentation,collision-induced dissociation(CID)is one of the most actively employed strategies for deriving sequence information of peptides[27,28].The combination of MS spectra and MS2fragment ion spectra was considered as the standard methodology for elucidating peptide sequences.
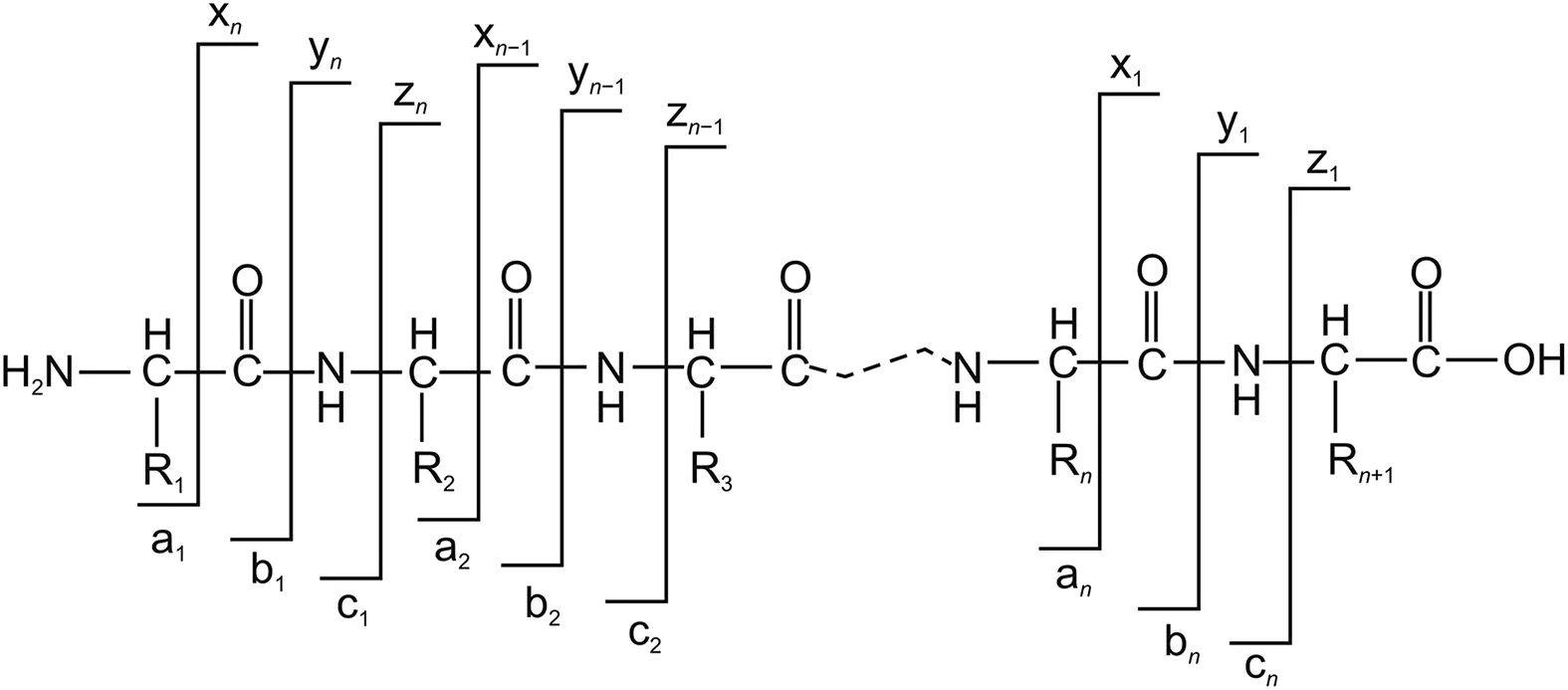
Fig.3.Schematic representation of established nomenclature used in the interpretation of product ions in the MS2spectra of peptides and proteins.
The schematic representation of the established nomenclature for the fragmentation pattern of a peptide and protein is depicted in Fig.3.A peptide can produce six types of fragment ions when cleaved.When the charge is retained by the N-terminal,the fragments are designated as an,bn,and cn,whereas when the charge is retained by the C-terminal,the fragments are assigned as xn,yn,and zn.The subscript n indicates the number of amino acid residues,and the superscript n denotes the charge of each fragment ion.Ions b and y are the most common complementary ions generated on peptide fragmentation.
To investigate the metabolic stability profiles of PEP06,the results of the first step of an extracted ion chromatogram(EIC)of different biological matrices using HRMS are shown in Fig.4.To further elucidate the metabolite structural information,fragment ion spectra of the parent peptide and corresponding metabolites were acquired by MS2spectrum.After incubating PEP06 in different matrices,43 metabolites were detected;Tables S1 and S2 enlist the peptide sequences,retention times,metabolite names,and observed metabolites in each tissue.
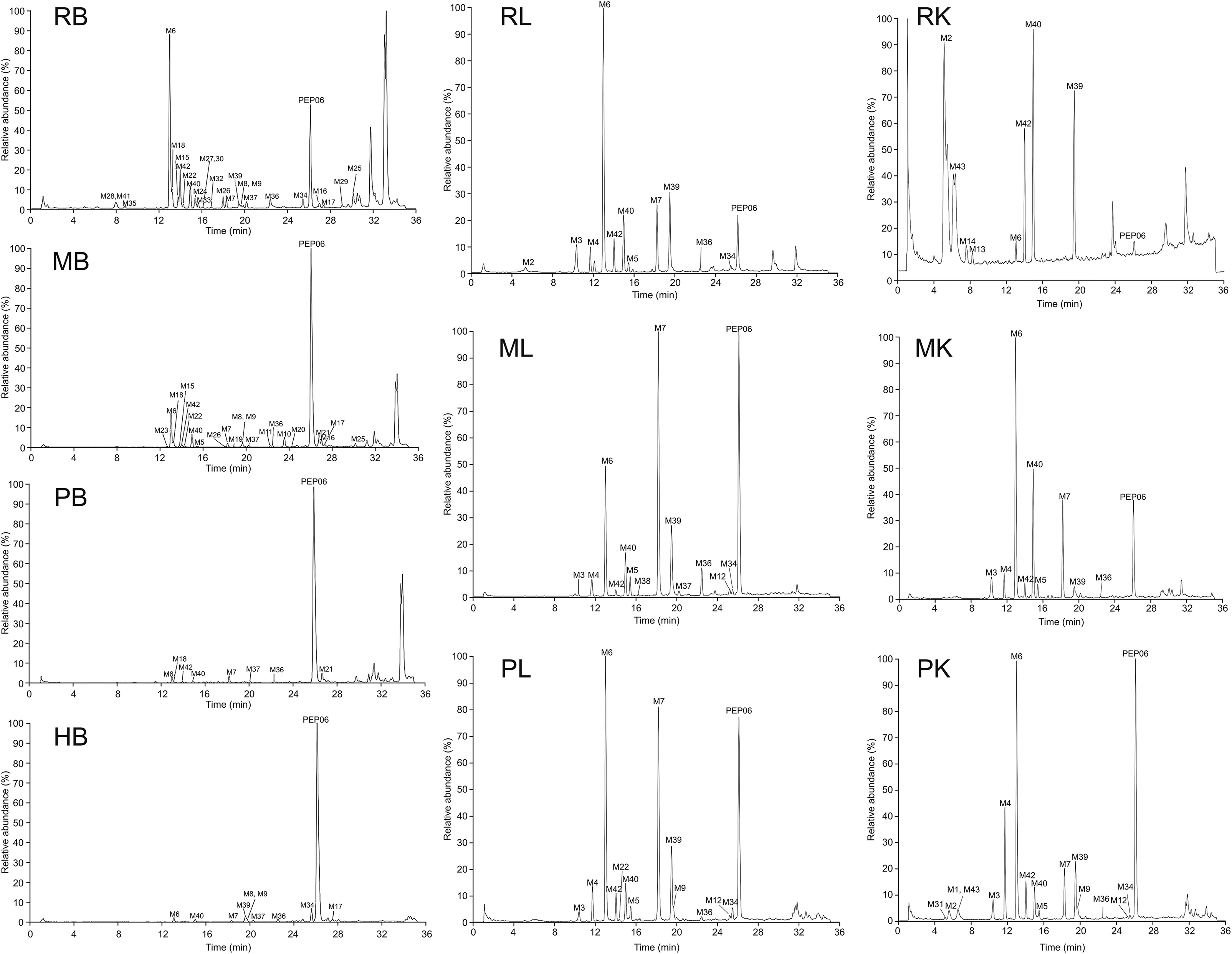
Fig.4.Extracted ion chromatograms from samples derived from different matrices of different species using high-resolution mass spectrometry(HRMS).RB:Sprague Dawley(SD)rats blood;MB:Cynomolgus monkeys blood;PB:Bama miniature pigs blood;HB:human blood;RL:SD rats liver;ML:Cynomolgus monkeys liver;PL:Bama miniature pigs liver;RK:SD rats kidney;MK:Cynomolgus monkeys kidney;PB:Bama miniature pigs kidney.
PEP06,which was artificially amidated by NH2to the C-terminal,possesses a monoisotopic accurate mass of 3,298.6433([M+H]+)and represents the charged states 1,100.2193[M+3H]3+,825.4163 [M+4H]4+,660.5345 [M+5H]5+,and 550.6133[M+6H]6+in the full scan of the MS spectrum.The most prominent peak in the full MS spectrum had a[M+5H]5+value of 660.73505,which was inconsistent with the theoretical value;the theoretical value reflected the value of the12C isotope.For small peptides,the theoretical value typically represents the most intense peak;however,for larger peptides,the12C isotope does not have the highest isotopic distribution.The CID spectrum of PEP06 confirmed that ion products were chiefly b and y ions.MS2spectrum of the fifth-charged ion(m/z 660.73505)generated a series of ions,and the interpretation for the product ions b6,b13,b14,b15,b16,b17,b18,b19,b21,b23,b24,b26,b27,b28,and b29was consistent with that of the parent peptide.Fig.5A shows the fullscan MS spectrum,and Fig.5B shows MS2spectrum supported by the structural assignment of various fragment ions.The mass spectra were confirmed by full mass determination and identification of product ions by MS2.
As shown in Tables S1 and S2,all the corresponding metabolites were cleaved from the peptide backbone although metabolites resulting from proteolytic hydrolysis were cleaved at both C and N termini.The names of metabolites listed in this research are designated by peptide sequence with respect to the parent compound.Metabolites M6(Fig.6A)and M40(Fig.6B)were observed in all samples incubated in the matrices,and the cleavage sites were His18-Leu19 and Leu22-Asn23,respectively.The semi-quantitative determination of metabolites abundance was calculated by comparing their peak area with that of the original parent compound.M6 was considered to be the most abundant metabolite,as seen from Table S3.The MS2spectra of all metabolites with structural assignments are shown in Table S4.
3.4.Further verification of the identity of metabolites
The identity of the corresponding metabolites was confirmed via experiments wherein the reference standard of synthesized metabolites was validated.Compared with observed samples in different matrices,the spiked samples of reference standard metabolites had the same retention time,similar full MS spectrum and MS2spectrum information.Upon further analysis,the amino acid sequences of M6 and M40 were further confirmed by analyzing the reference standard metabolites of RGDRGDMHSHRDFQPVLH and NSPLSGGM-amidated(artificially amidated by NH2to the C-terminal)based on HRMS.The cleavage sites of other metabolites represented were observed between residues M7(Leu19-Val20),M16(Arg1-Gly2),M17(Gly2-Asp3),M21(Asp3-Arg4),M25(His10-Arg11),M33(His18-Leu19,Asn23-Ser24),M34(His18-Leu19),M39(Ala21-Leu22),and M42(Asn23-Ser24).The above-mentioned metabolites were synthesized and compared with samples in retention time,full MS spectrum and MS2spectrum information for further confirmation.
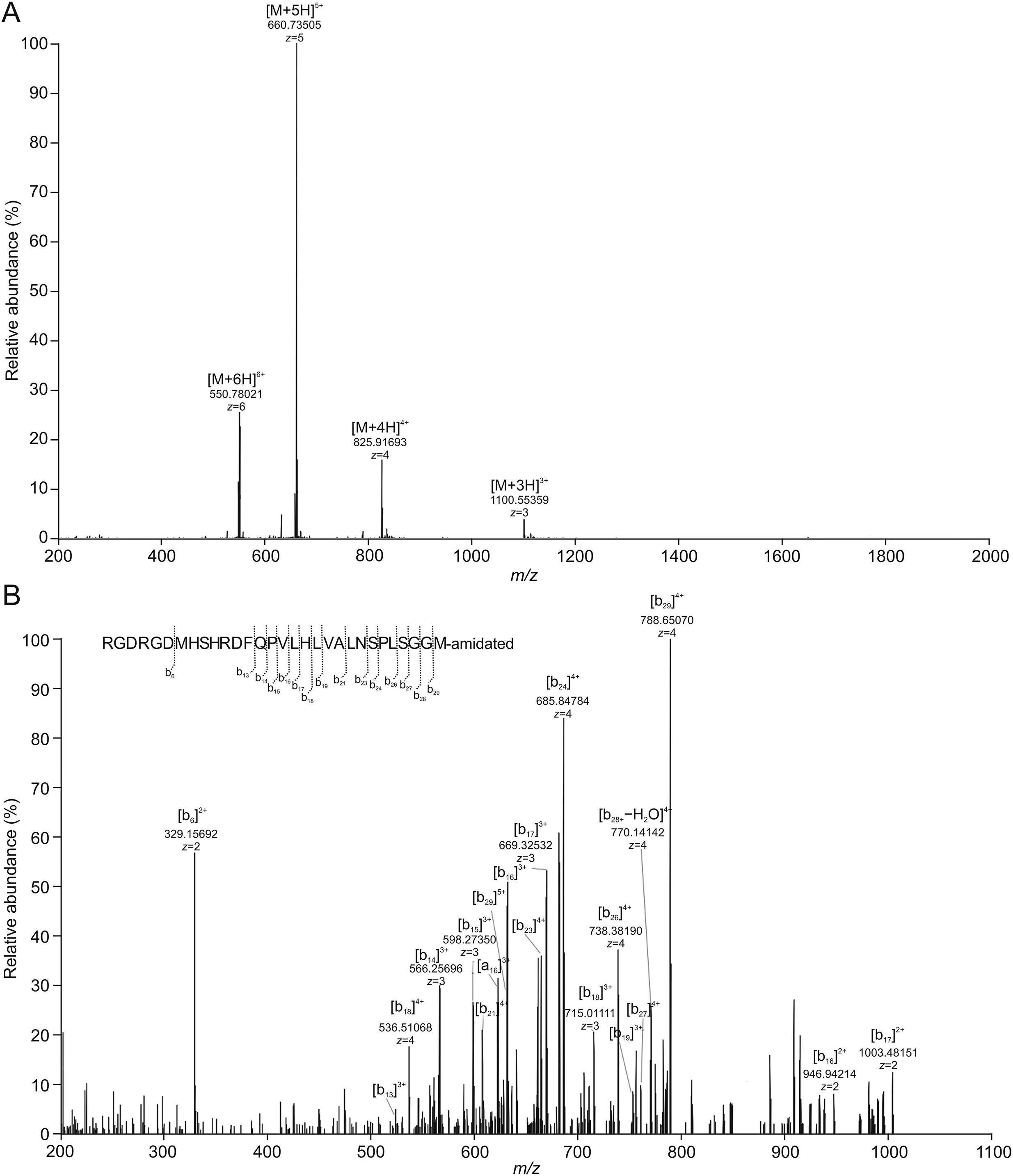
Fig.5.Identification of PEP06 by high-resolution mass spectrometry(HRMS)in vitro.(A)Full-scan MS spectrum of PEP06 analyzed by liquid chromatography-mass spectrometry(LC-MS).(B)MS2spectrum of PEP06 analyzed by LC-MS.
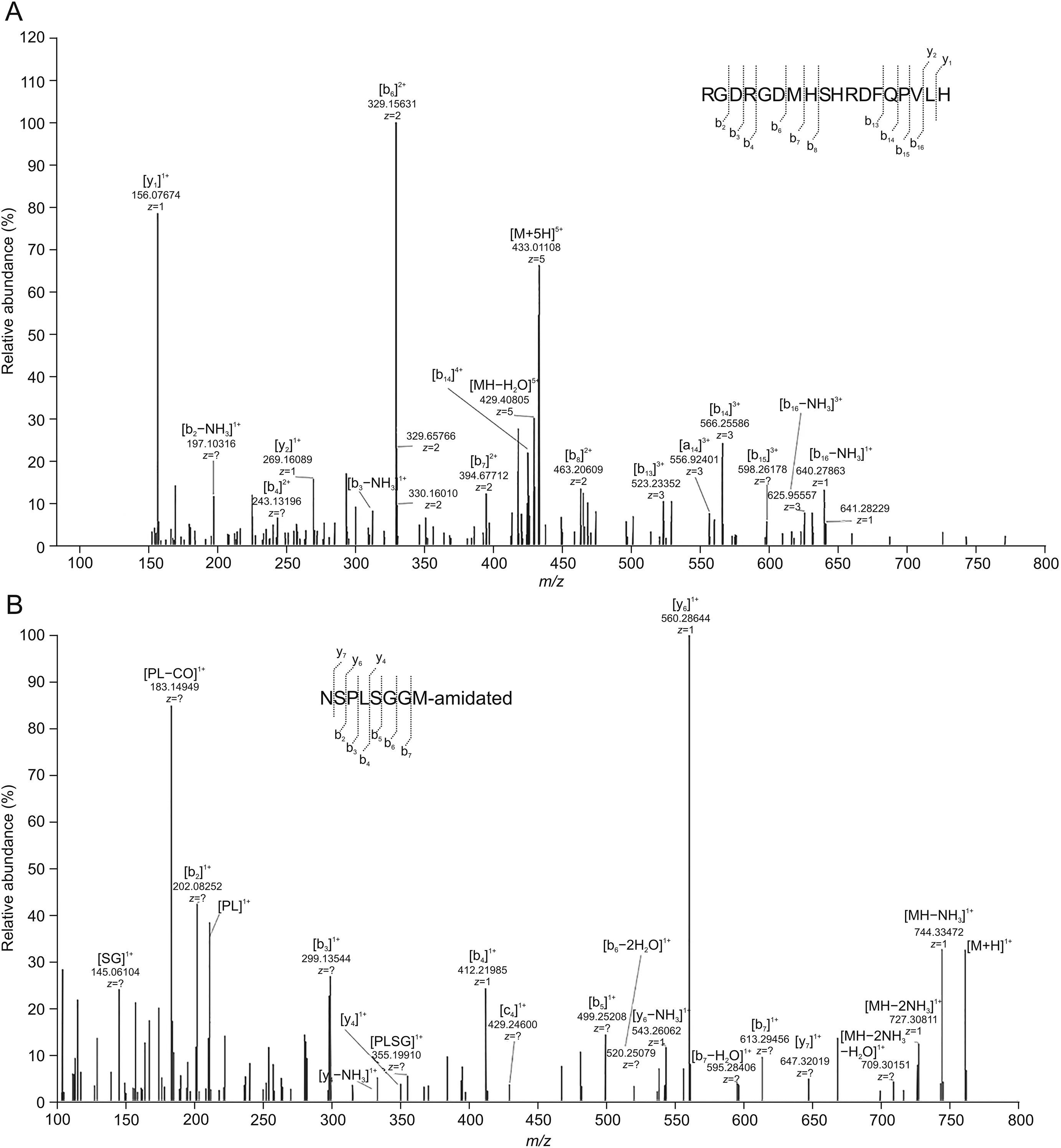
Fig.6.Identification of metabolites M6 and M40 by high-resolution mass spectrometry(HRMS)in vitro.(A)MS2spectrum of M6.(B)MS2spectrum of M40.
4.Conclusion
PEP06,a novel endostatin-RGDRGD 30-amino-acid polypeptide with antitumoral properties,was synthesized by terminally fusing the RGDRGD hexapeptide at the N terminus of the active endostatin fragment.Moreover,PEP06 has demonstrated potent antitumor effects both in vitro and in vivo in previous studies[29].Most importantly,PEP06 has lived up to our expectations with respect to tumor treatment and is undergoing phase I clinical trials in China.Such clinical research supports its further development as a new and effective antitumor drug for use in the future.
In summary,our in vitro data show that PEP06 underwent rapid enzymatic degradation in the different matrices of living cells obtained from different animal species.The results confirmed that blood and the liver and kidneys play important roles in the metabolism and clearance of molecular degradation peptides from PEP06.We optimized the LC and HRMS methods for acquiring and characterizing metabolites,which is paramount for achieving an ideal response,acceptable peak shape,and sensitivity necessary to identify peptide fragments and metabolites.In the present study,the PEP06 metabolites were analyzed in vitro by MS,and a total of 43 metabolites were identified.The structures of 11 of these metabolites(M6,M7,M16,M17,M21,M25,M33,M34,M39,M40,and M42)were further confirmed by comparing their chromatographic characteristics,full-scan MS spectra,and MS/MS spectra with those of reference standards of synthesized metabolites.This approach to the discovery of metabolites from peptide therapeutics demonstrates a potential value for developing bioactive peptide metabolites as antitumor drugs.Furthermore,future pharmacology research on these bioactive downstream metabolites of PEP06 uncovered in this study can go a long way in treating tumors.
CRediT author statement
Liyun Niu:Conceptualization,Methodology,Software,Validation,Formal analysis,Investigation,Data curation,Writing-Original draft preparation,Reviewing and Editing,Visualization;Huiyu Zhou:Software,Investigation;Yueru Lian:Investigation;Ya Gao:Investigation;Yulu Liu:Investigation;Ruolan Gu:Resources,Supervision;Zhuona Wu:Resources;Xiaoxia Zhu:Resources,Supervision;Hui Gan:Resources;Zhiyun Meng:Conceptualization,Funding acquisition;Guifang Dou:Conceptualization,Funding acquisition.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Appendix A.Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jpha.2022.03.002.
 Journal of Pharmaceutical Analysis2022年5期
Journal of Pharmaceutical Analysis2022年5期
- Journal of Pharmaceutical Analysis的其它文章
- Caenorhabditis elegans deep lipidome profiling by using integrative mass spectrometry acquisitions reveals significantly altered lipid networks
- Effective extraction of fluoroquinolones from water using facile modified plant fibers
- Rapid fabrication of zwitterionic sulfobetaine vinylimidazole-based monoliths via photoinitiated copolymerization for hydrophilic interaction chromatography
- Effects of polyol excipient stability during storage and use on the quality of biopharmaceutical formulations
- Multimodal integrated strategy for the discovery and identification of quality markers in traditional Chinese medicine
- The dynamic metabolic profile of Qi-Yu-San-Long decoction in rat urine using UPLC-QTOF-MSEcoupled with a post-targeted screening strategy
