Long-term and non-invasive in vivo tracking of DiD dye-labeled human hepatic progenitors in chronic liver disease models
Chaturvedula Tripura,Srinivas Gunda, Sandeep Kumar Vishwakarma, Avinash Raj Thatipalli, Jedy Jose,Mahesh Kumar Jerald,Aleem Ahmed Khan,Gopal Pande
Abstract
BACKGROUND
Chronic liver diseases (CLD) are the major public health burden due to the continuous increasing rate of global morbidity and mortality. The inherent limitations of organ transplantation have led to the development of stem cell-based therapy as a supportive and promising therapeutic option. However, identifying the fate of transplanted cells in vivo represents a crucial obstacle.
AIM
To evaluate the potential applicability of DiD dye as a cell labeling agent for longterm, and non-invasive in vivo tracking of transplanted cells in the liver.
METHODS
Magnetically sorted, epithelial cell adhesion molecule positive (1 × 106 cells/mL) fetal hepatic progenitor cells were labeled with DiD dye and transplanted into the livers of CLD-severe combined immunodeficiency (SCID) mice. Near-infrared (NIR) imaging was performed for in vivo tracking of the DiD-labeled transplanted cells along with colocalization of hepatic markers for up to 80 d. The existence of human cells within mouse livers was identified using Alu polymerase chain reaction and sequencing.
RESULTS
NIR fluorescence imaging of CLD-SCID mice showed a positive fluorescence signal of DiD at days 7, 15, 30, 45, 60, and 80 post-transplantation. Furthermore, positive staining of cytokeratin, c-Met, and albumin colocalizing with DiD fluorescence clearly demonstrated that the fluorescent signal of hepatic markers emerged from the DiD-labeled transplanted cells. Recovery of liver function was also observed with serum levels of glutamic-oxaloacetic transaminase, glutamate-pyruvate transaminase, and bilirubin. The detection of human-specific Alu sequence from the transplanted mouse livers provided evidence for the survival of transplanted cells at day 80.
CONCLUSION
DiD-labeling is promising for long-term and non-invasive in vivo cell tracking, and understanding the regenerative mechanisms incurred by the transplanted cells.
Key Words: Chronic liver diseases; Cell transplantation; Cell tracking and imaging; DiD; Hepatic progenitors
INTRODUCTION
Chronic liver diseases (CLD) represent one of the leading causes of morbidity and mortality worldwide, specifically in developing countries. They result from the progressive deterioration of liver functions which is caused by the continuous process of inflammation, destruction, and inadequate repair of the liver parenchyma leading to cirrhosis. Liver cirrhosis is characterized by irreversible distortion of the liver architecture in the form of fibrosis, scar formation, the occurrence of several regenerative nodules, vascular reorganization, and immense liver failure. The major aim of the current treatment approaches is to halt the progression of CLD into more severe forms, and further reduce the associated complications representing the need for an appropriate multidisciplinary approach. The clinical signs and symptoms of CLD can be nonspecific; hence the major management perceptions are the elimination of underlying causes, management of portal hypertension, and personalized therapy for each associated condition. Although such strategies provide temporary support to the failing liver; they cannot prevent long-term disease progression. Hence, more effective management approaches are required to overcome such hurdles and bridge the gap of a long-term therapeutic window to improve health-related quality of life.
Cell therapy is an emerging technology and has shown significant promise in addressing the current demand for alternative options to liver transplantation to improve liver functions and act as a supportive bridge therapy in CLD[1,2]. However, it is necessary to ascertain appropriate cell types from widely acceptable sources and resolve several unanswered questions before utilizing cell-based technology in the clinical setting. Since the last two decades, transplantation of different types of cells from various tissue sources such as autologous bone marrow-derived mononuclear cells and mesenchymal stem cells (MSCs), and allogenic fetal hepatic progenitor cells (fHPCs) have shown promising outcomes in clinical studies of CLD[3-7]. In addition, several preclinical studies have also demonstrated the potential applicability of such approaches in immune-compromised/deficient mice to improve the current understanding of the safety, efficacy, and functionality of human stem/progenitor cells post-transplantation[8-11]. While several of these studies have proved the safety and involvement of the transplanted cells in liver recovery, the availability of methods for easy and long-term tracking of infused cells would be extremely beneficial in determining their viability, bio-distribution, homing, and differentiation which represents a major roadblock for cell-based therapies in clinical settings.
The majority of existing strategies employ radioisotopes, magnetic particles, fluorescent tags, or reporter genes as cell labeling agents prior to transplantation in preclinical settings[12]. Furthermore, non-invasive radionuclide imaging methods such as single-photon emission tomography and positron emission tomography using radionuclides [Technetium (99mTc) and111In-oxine] are currently employed in clinical settings[4,6,13]. However, the short life of radionuclides limits their wider applicability due to monitoring of the immediate cellular behavior for only a few hours. Magnetic resonance imaging (MRI) is another non-invasive imaging method that has been explored for cell tracing in preclinical CLD models for up to 1-2 wk[14,15]. While MRI offers good spatial resolution and contrast, it is less sensitive and is not effective for follow-up studies due to the gradual loss of signal intensity[12]. Hence, several other tracking methods based on reporter gene expressions, such as fluorescence imaging and bioluminescence imaging are being successfully employed for monitoring the fate of transplanted cells in animal models of liver injury[16,17]. Although this method enables long-term cell tracking, the safety concerns owing to genetic manipulation represent a major hurdle for clinical translation. Hence, direct labeling of the cells without involving genetic manipulation represents a crucial need for a sensitive, relatively safer, and less cumbersome process for tracking transplanted cells in both preclinical and clinical settings.
In the present study, long-term and non-invasive tracking of transplanted fHPCs was evaluated in an experimental severe combined immunodeficiency (SCID) mouse model of CLD using DiD. DiD is a carbocyanine dye having good photochemical properties of strong fluorescence, and stability[18-20]. It is a cationic dye and belongs to the family of lipid intercalating long alkyl side-chain carbocyanine derivatives that have a long-range (540-780 nm) emission. Due to the long-range emission, tissue autofluorescence of DiD is minimum, permitting the use of other fluorochromes such as fluorescein isothiocyanate (FITC) for co-localization studies to evaluate the expression of other essential markers specific to transplanted cells in the recipient tissue. Moreover, the process of labeling the cells using DiD is easy due to its excellent efficiency for integration and diffusion into the cell membranes[18,19,21,22]. Although DiD is insoluble in water, its fluorescence is readily detected when incorporated into the cell membranes. Therefore, it has been classified as one of the most appropriate carbocyanine families of dyes in cell labeling and tracking. After incorporation into cell membranes, it diffuses laterally within the plasma membranes, resulting in staining of the entire cell. Structural similarity with the cell membrane phospholipids and prolonged dye retention within the cells are among the major advantages of DiD for live organisms. Hence, DiD has been used for labeling different types of cells without interfering with cellular differentiation; however, the effects of DiD labeling on human liver cells and its effect on the in vivo retention of labeled human liver cells remain to be investigated.
We specifically utilized magnetically sorted fHPCs using epithelial cell adhesion molecule (EpCAM) as a surface marker due to its associated crucial functions such as cell-to-cell adhesion, proliferation, maintenance of pluripotent state, and regulation of differentiation and migration[23]. It has also been demonstrated that EpCAM-positive HPCs are highly proliferative and have diminished class II MHC presentation, and are classified as immature cells suitable for regenerative applications[24]. In our earlier study, EpCAM-positive fHPCs revealed a significant improvement in liver functions and increased disease-free life span in patients with end-stage CLD[6]. Thus, using EpCAM-positive fHPCs could highlight how good DiD is for long-term, non-invasive, and real-time monitoring of cell survival, and structural and functional improvements in preclinical models of CLD post-transplantation. Accordingly, the present study aimed to shed light on the fate of DiD-labeled human liver cells in CLDSCID mice using live imaging up to 80 d post-transplantation.
MATERIALS AND METHODS
Animals
Experimental animals were obtained from inbred colonies of SCID mice (strain: NOD.CB17-Prkdcscid/J) and maintained at the animal facility of the Centre for Cellular and Molecular Biology (CCMB), Hyderabad, India. This study was approved by the Institutional Animal Ethics Committee (Animal trial registration number 20/1999/CPCSEA dated 10/03/1999) of CCMB. All the animal experimental procedures were performed in accordance with the approved ethical guidelines of CCMB for the care and use of animals. All the animals were maintained in standard ventilated cages with a 12 h light-dark cycle and were fed ad libitum.
Development of the CLD mousemodel
CLD mouse model was generated using 25% carbon tetrachloride (CCl4, Rankem, India) diluted with mineral oil (Sigma, United States). A sub-lethal dose of diluted CCl4was administered according to 125 μL/kg body weight in each animal. A total of 26 mice (either sex) at eight weeks of age were randomly assigned to the vehicle control group (n = 4) and the CCl4group (n = 22). The CCl4group received intraperitoneal injections of diluted CCl4twice a week for 4 wk and the vehicle control mice received only mineral oil. After 4 wk, liver damage was confirmed by changes in liver enzymes and liver tissue histology. For the biochemical evaluation of liver damage, 100-150 μL of blood was collected by orbital sinus puncture. Serum levels of total bilirubin, glutamate-pyruvate transaminase (SGPT), and glutamicoxaloacetic transaminase (SGOT) were measured by the Jendrassik-Grof and Reitman-Frankel’s methods, respectively, using kits (Coral Clinical Systems, India). The mice after 4 wk of CCl4(referred to as CLD-SCID mice) were ready for cell transplantation (TX).
Isolation of human fetal hepatic progenitors
The total fetal liver cells (tFLCs) isolation protocol was approved by the Institutional Ethics Committee of Deccan College of Medical Sciences, Hyderabad. Informed consent was obtained prior to sample collection, and cell processing was performed according to the ethical guidelines for the use of human cells. Briefly, the whole liver was dissected from spontaneously aborted fetuses (n = 3, 10-12 wk gestation), and perfused twice with ice-cold phosphate-buffered saline (PBS) for 5 min to eliminate circulating peripheral blood cells, followed by digestion with 0.025% collagenase in 1× PBS for 5 min at room temperature. Then the liver tissue was minced with a scalpel blade and disintegrated into a singlecell suspension by passing through 40 μm cell strainers (BD Biosciences, United States). Cell viability and counting were performed using the trypan blue dye exclusion test.
Flow cytometry
For flow cytometry analysis, a single cell suspension of 2 × 106tFLCs was fixed in 4% paraformaldehyde for 15 min at room temperature. Following fixation, the cells were washed twice with 1× PBS and stained with anti-human EpCAM (CD326) antibody conjugated with FITC (Miltenyi Biotech, Germany) for 30 min. Cells were washed once with 1× PBS before analyzing on FACS CaliburTM(BD Biosciences, United States) using 488 nm argon laser emission at 530/30 BP filter. The data were analyzed and plotted using Kaluza software 1.5a (Beckman Coulter Inc., United States).
Enrichment of EpCAM-positive fHPCs from tFLCs
To isolate EpCAM-positive cells, a 5 × 107tFLC suspension in 500 μL buffer containing FCR blocking reagent was incubated with anti-human EpCAM antibody (Miltenyi Biotech, Germany) conjugated with magnetic beads at 4°C for 30 min and sorted using the magnetic cell sorter, AutoMACS according to the manufacturer’s instructions (Miltenyi Biotech, Germany). Magnetically activated cell sorting (MACS) enriched EpCAM-positive cells were collected and resuspended in RPMI 1640 medium (Sigma, United States) supplemented with 10% fetal bovine serum (FBS, Sigma, United States). Cell isolation and MACS sorting procedures were carried out under sterile conditions in a Class 100 biosafety cabinet.
Immunocytochemistry
MACS-sorted EpCAM-positive cells were fixed in 4% paraformaldehyde, and cytospin preparations were conducted on “Probe-on-Plus” slides (Fisher Scientific, United States). Cells were blocked with 10% goat serum, and stained with mouse monoclonal anti-human EpCAM antibodies directly conjugated with FITC (Miltenyi Biotech, Germany), and co-stained with either anti-cytokeratin (CK) 8+18+19 or anti-c-Met (Abcam Inc., MA, United States) primary antibodies. Alexa 594 (Molecular Probes, United States) was used as the secondary antibody. Images were captured using a confocal laser scanning microscope (Leica, Germany, SP2 AOBS).
DiD-labeling and intra-hepatic cell TX in SCID-CLD mice
MACS sorted EpCAM-positive cells (1 × 106cells/mL) in Hank’s buffered salt solution (HBSS) were labeled with DiD dye by adding 5 μL of DiD cell labeling solution (Life Technologies, Eugene, United States) to the cell suspension and incubating for 20 min at 37°C. DiD-labeled cells were washed thrice with HBSS, and resuspended in the same buffer. 1 × 105cells (100 μL) were injected directly into the liver lobes of the CLD mice (n = 14) at a single site using a 26-gauge needle. CLD mice receiving plain HBSS buffer served as non-transplanted (non-TX) controls (n = 5). Post-transplantation, mice were maintained in different cages, imaged, and sacrificed at different time points.
Long-term in vivo and ex vivo imaging
DiD-specific fluorescence from the transplanted animals was detected on the Multispectral FXPRO Fluorescence Imager (Carestream-KODAK, United States) using 630 nm excitation and 670/30 BP emission filters. Highly sensitive fluorescence images were combined with the high resolution X-ray images to precisely locate the DiD-labeled cells. Animals were imaged prior to TX (0 d) and after 7, 15, 30, 45, 60, and 80 d, post-TX. For each imaging experiment, the animals were anaesthetized with Xylazine (5 mg/kg body weight) and Ketamine (25 mg/kg body weight), the abdominal hair was shaved, placed within the chamber, and imaged. After in vivo imaging at days 15, 30, and 80 post-TX, mice were sacrificed, liver tissues were excised, and imaged (ex vivo imaging) to locate the fluorescing lobe for further processing. The fluorescing area of the liver lobe at day 15 and day 80 was immersed in OCT mounting solution and stored at -80°C. Liver lobes of the non-TX mice were randomly selected and processed similarly. The liver lobes at day 30 post-TX were processed for paraffin embedding and histology.
Histology analysis
The excised liver lobes were fixed in 10% buffered formalin and processed for paraffin embedding. Paraffin-embedded liver tissues were sectioned at 4.0 μm thickness using a rotatory microtome (Leica RM2135, Germany), and stained with Hematoxylin and Eosin, and Sirius Red (Sigma-Aldrich, United States) using standard protocols. Bright-field images were acquired from both control and CLD mice (n = 3) at 10× and 40× using an Olympus inverted microscope (IX3-SSU, Tokyo, Japan). A total of 20-25 random fields captured with the 10× objective was used to calculate the total collagen area using Image J (1.5 2q) software, and expressed as total collagen percent area (% CPA).
Immunostaining
Serial cryosections (7.0 μm thick) of transplanted and non-transplanted mouse liver lobes at 15 and 80 d were stained to determine the expression of hepatic markers using anti-CK, anti-c-Met, and anti-human albumin (MP Biomedicals) primary antibodies, and detected with Alexa 488 (Molecular Probes, United States) secondary antibody. Images were captured using either a confocal laser scanning microscope (Leica, Germany, SP2 AOBS), or the Axioimager Z2 Fluorescence microscope (Zeiss, Germany).
Alu sequence analysis
DNA was isolated from the fHPCs prior to transplantation and from the liver tissues of transplanted and non-transplanted mice at day 80. Isolated DNA was analyzed using human-specific primers for Alu sequence (Forward primer 5’-GGCGCGGTGGCTCACG-3’, Reverse primer 5’-TTTTTTGAGACGGAGTCTCGCTC-3’). Polymerase chain reaction (PCR) was performed in a 25 μL reaction mixture containing 1 μL DNA, 2.5 μL 10× complete PCR Buffer with MgCl2, 1 μL of 10 mmol/L dNTPs, 0.5 μL forward and 0.5 μL reverse primers, and 0.2 μL Taq DNA Polymerase (5 U/mL) with an initial denaturation step of 94°C for 5 min, followed by 35 cycles of a three-step program of 94°C for 30 s, 54°C for 30 s and 72°C for 45 s, followed by a final extension step at 72°C for 5 min. The PCR products were electrophoresed on a 2% agarose gel and observed under UV with ethidium bromide staining. The images were captured using the Gel Documentation System (BIO-RAD, United States). The PCR product was cleaned using QIAquick Gel Extraction Kit (Qiagen, United States), and sequenced. The amplicon sequences were analyzed using the ClustalW2 online tool (https://www.ebi.ac.uk/Tools/msa/clustalw2/).
Statistical analysis
All statistical analyses were performed using GraphPad Prism software (ver. 5.0). Data are presented as mean ± standard error of the mean. Paired Student t-test at 95% confidence interval for a P value of ≤ 0.05 was considered statistically significant.
RESULTS
The schematic representation in Figure 1 shows the different steps involved in measuring different outcomes throughout the study process. Isolated tFLCs were enriched for EpCAM-positive cells by MACS in step 1 (Figure 1A), labeled with DiD dye in step 2 (Figure 1B), and then transplanted into CLD mouse livers (Figure 1C). Furthermore, live non-invasive near-infrared (NIR) imaging was performed at regular intervals using a small animal imaging system to detect the fluorescence signal (Figure 1D). However, ex vivo imaging of the excised liver was performed to confirm the localization of the fluorescence in mice liver post-TX (Figure 1E).
Immunophenotyping of tFLCs to identify the proportion of fHPCs
Human tFLCs stained with FITC-tagged anti-CD326 (EpCAM) antibodies were analyzed using flow cytometry. The gating strategy and the analysis of cell fluorescence vs cell size, and the overlaid histogram of the unstained and EpCAM-stained cells were acquired (Figure 2A and B). Of these tFLCs, 49 ± 23% of cells were found to be EpCAM positive and designated as fHPCs.
NIR imaging of DiD-labeled EpCAM-positive cells in vitro
MACS-sorted EpCAM-positive fHPCs were characterized for the expression of specific hepatic cell markers such as CK and c-Met together with EpCAM (Figure 2C). Magnetically sorted EpCAM-positive cells which were labeled with DiD dye showed a separate peak corresponding to the DiD fluorescence (Figure 2D). Furthermore, before transplantation, the DiD-labeled EpCAM-positive cells in the tube were visualized for fluorescence in the multispectral imaging system. The overlay image of the NIR fluorescence and X-ray showed a positive signal only in the tube containing DiD-labeled cells, while the tube with non-labeled cells was devoid of any such fluorescence (Figure 2E).

Figure 1 Schematic presentation of the study. A: Total fetal liver cells were enriched for EpCAM-positive cells by magnetically activated cell sorting; B: Cells were labeled with DiD and visualized in the tube by near-infrared imaging as red fluorescence; C: Cells were intra-hepatically transplanted into injured livers of severe combined immunodeficiency mice; D: In vivo imaging of live animals was performed at specified time points; E: Ex vivo imaging of liver confirmed the retention of DiD-labeled transplanted cells in the liver. EpCAM: Epithelial cell adhesion molecule; SCID: Severe combined immunodeficiency; MACS: Magnetic activated cell sorting.
Long-term tracking of DiD-labeled cells post-TX in CLD-SCID mice
Assessment of the biochemical and histological changes in CLD-SCID mice showed an increase in serum parameters (SGOT, SGPT, and bilirubin), and collagen accumulation compared to the control animals, confirming liver injury after 4 wk of CCl4injection (Supplementary Figure 1). These CLD mice were utilized for cell transplantation (at day 0), and tracking through in vivo imaging on different days from day 0 to day 80 (Figure 3A). NIR fluorescence imaging of CLD-SCID mice before intra-hepatic transplantation of DiD-labeled fHPCs cells at day 0 did not show fluorescence, while mice at days 7, 15, 30, 45, 60, and 80 post-TX showed a positive fluorescence signal of DiD (Figure 3B). The overlay images showed DiD fluorescence in the upper part of the abdominal cavity near the rib cage, suggesting that the cells continue to localize in the liver lobes until day 80 post-TX. The non-TX mouse abdomen lacked such fluorescence signals (Figure 3C). To further confirm the localization, the liver was excised on day 15 and day 80 post-TX, and ex vivo imaging was performed which confirmed the localization of DiD-labeled cells within the mice livers (Figure 3D). These results indicated that DiD-labeled cells can be efficiently visualized and tracked for a longer duration post-TX both in vivo and ex vivo.

Figure 2 Flow cytometry characterization of epithelial cell adhesion molecule expressing fetal hepatic progenitors, and DiD labeling of magnetically activated cell sorting enriched epithelial cell adhesion molecule-positive cells. A and B: Total fetal liver progenitors were stained with epithelial cell adhesion molecule (EpCAM)-fluorescein isothiocyanate (FITC) antibody and the representative dot plots of (A) unstained and EpCAM (CD326) stained cells and (B) overlay histogram; C: Colocalization of pan CK and c-Met with EpCAM-FITC expression (Scale bar = 10 μm); D: Overlay histogram of magnetically sorted EpCAM-positive cells labeled with DiD; E: Near-infrared fluorescence imaging and X-ray overlay image of DiD-labeled and unlabeled EpCAM-positive cells in the tube. EpCAM: Epithelial cell adhesion molecule; NIR: Near-infrared; DAPI: 4',6-diamidino-2-phenylindole; MACS: Magneticallyactivated cell sorting.
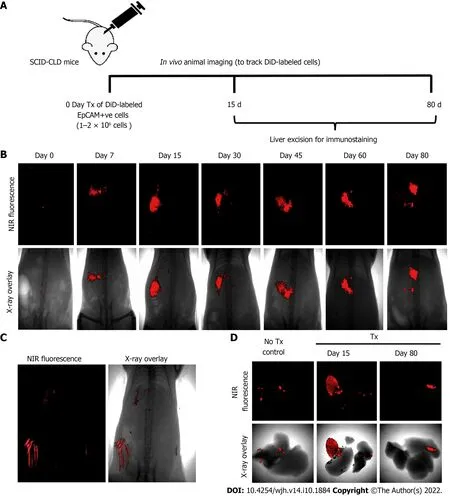
Figure 3 Long-term in vivo tracking of transplanted cells. A: Schematic representation showing the timeline for cell transplantation in mice with chronic liver diseases, imaging, and animal sacrifice; B: In vivo near-infrared (NIR)-fluorescence images (top panel) overlay with X-ray images (lower panel) of mice posttransplantation at indicated time periods; C: NIR-fluorescence image overlay with X-ray image of non-transplanted control mice (hair autofluorescence can be seen);D: Ex vivo imaging of excised livers of non-transplant control mice and transplanted mice at 15 and 80 d post-transplantation.
Co-expression of hepatic markers with DiD in the transplanted mouse livers
To evaluate the expression of hepatic markers in transplanted cells, frozen mouse liver sections were obtained from the portion of the liver that displayed a DiD-positive fluorescence signal at day 15 and day 80 post-TX. Serial sections of the liver transplanted with DiD-labeled fHPCs and non-transplanted control mice were obtained and stained for CK and c-Met (Figures 4 and 5). The presence of DiD fluorescence only in the transplanted mice confirmed the existence of transplanted cells, while nontransplanted mice did not show DiD fluorescence in liver tissue sections. In addition, positive staining for both CK (Figures 4A, 5A and 5C) and c-Met (Figures 4B, 5B and 5D) colocalization with DiD fluorescence clearly demonstrated that the fluorescent signal of the hepatic markers, CK and c-Met emerged from the DiD-labeled transplanted cells.
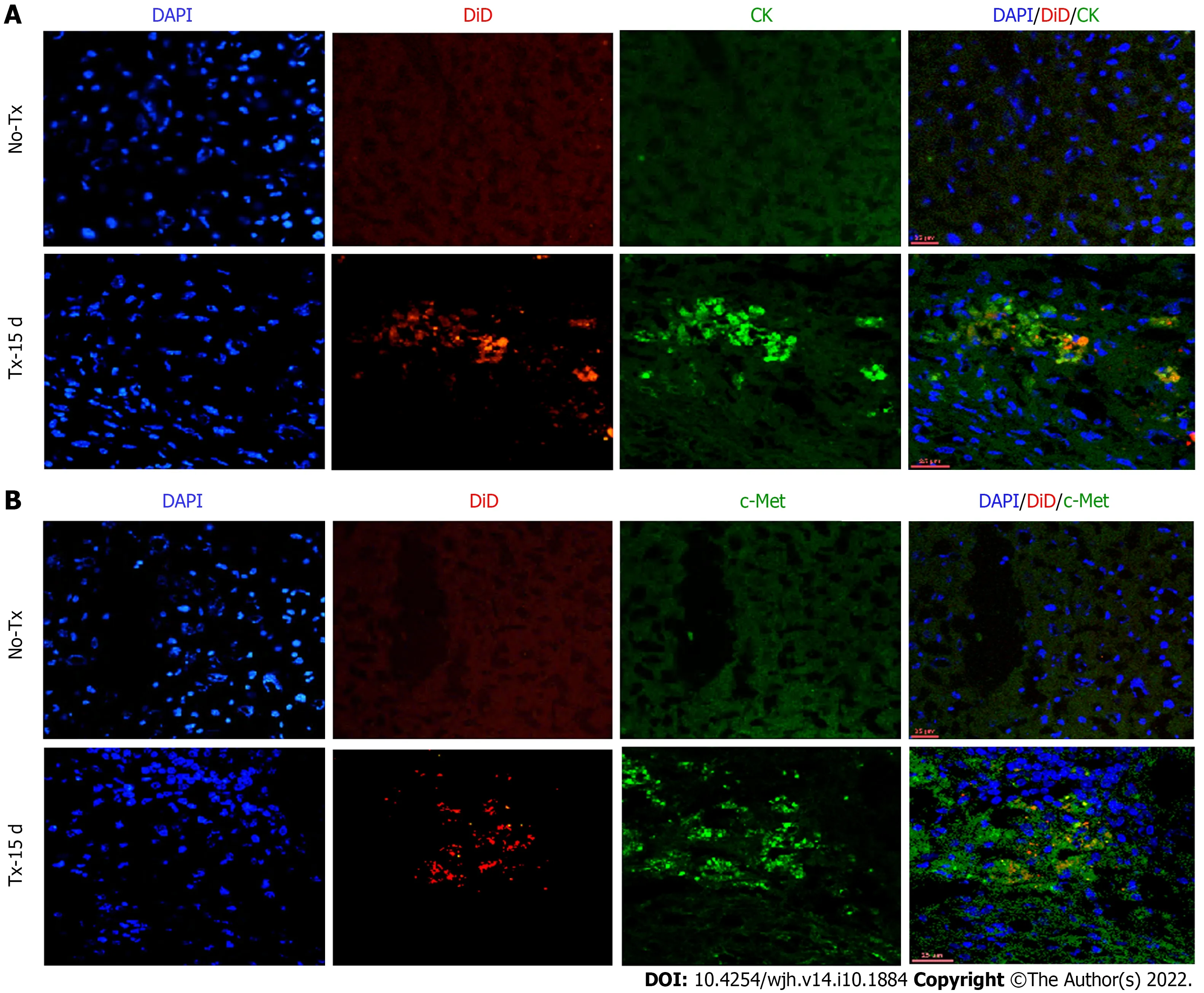
Figure 4 Co-expression of hepatic markers in the transplanted livers 15 d post-transplantation. Confocal images of liver tissue cryosections of transplanted (TX-15 d) and non-transplanted (No-TX) mice showing expression of CK and c-Met colocalization with DiD-labeling only in the transplanted mice. The non-transplanted mouse livers were negative for CK, c-Met and DiD (Scale bar = 25 μm). A: CK: cytokeratin; B: c-Met; DAPI: 4',6-diamidino-2-phenylindole.
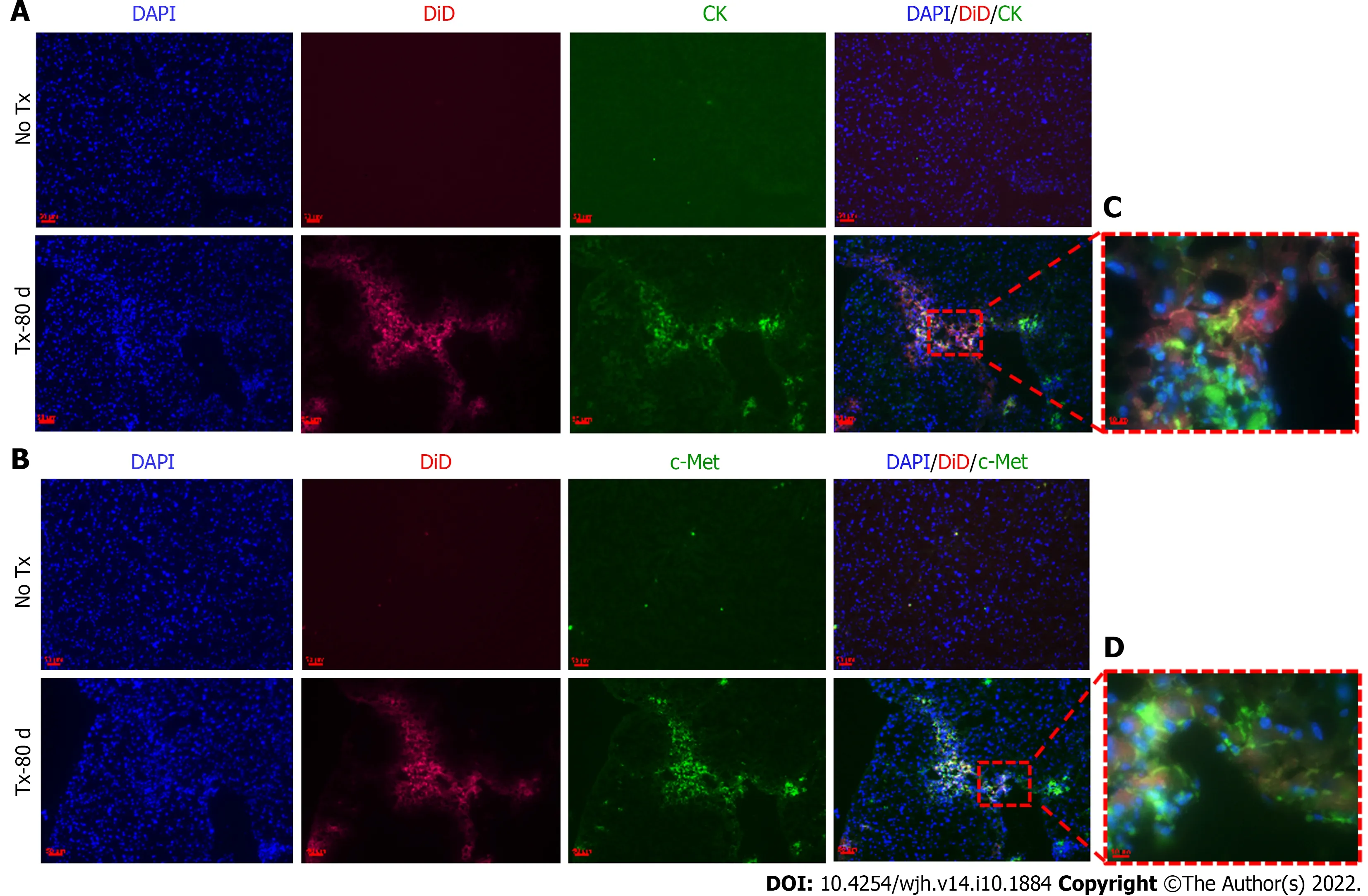
Figure 5 Detection of hepatic markers in transplanted mouse livers 80 d post-transplantation. A and B: Fluorescent microscopic images of mouse liver tissue cryosections that continue to show DiD signals 80 d post-transplantation and show co-expression of CK and c-Met only in the transplanted (TX-80 d)mouse livers, but not in the non-transplanted (No-TX) livers (scale bar = 50 μm); C and D: 63 × images showing the colocalization of CK and c-Met staining with DiD labeling, respectively (Scale bar = 50 μm and 10 μm). CK: cytokeratin; DAPI: 4',6-diamidino-2-phenylindole.
Tracing the effect of transplanted DiD-labeled cells through improved liver function parameters
After 15 d of transplantation, recovery of mouse liver function was analyzed by assaying liver enzymes SGOT, SGPT, and bilirubin. Serum SGOT and SGPT levels were significantly reduced in the transplanted mice compared to the non-transplanted mice (Figure 6A), while serum bilirubin was reduced to normal levels in both groups. Thus, the serum enzyme parameters suggested improved liver function in the transplanted mice compared to the non-transplanted mice. Furthermore, the lower collagen percentage area in transplanted mice compared to the non-transplanted mice suggested that this recovery may be attributed to the transplanted fHPCs (Figure 6B and C).

Figure 6 Liver enzyme and histology assessment of improved liver function parameters. A: Analysis of serum bilirubin, serum glutamic-oxaloacetic transaminase, (SGOT) and serum glutamate-pyruvate transaminase (SGPT) measured 15 d post-transplantation (TX) and compared to the non-transplanted (No-TX)mice (n = 4); B and C: Hematoxylin and Eosin (H&E) and Sirius Red staining of liver tissue sections of non-transplanted and transplanted mouse liver tissue at 30 d post-transplantation showed a significant decrease in collagen percent area in the transplanted mouse livers as compared to the non-transplanted mouse livers (aP <0.001). H&E: Hematoxylin and Eosin; SR: Sirius Red.
The above assumption was further confirmed by the positive expression of hu-ALB in CLD-SCID mouse liver cells at day 80 post-TX (Figure 7). ALB-positive cells were also tested for their colocalization with DiD to confirm the effect was only in labeled cells. The non-transplanted mouse livers tested negative for ALB staining. The presence of hu-ALB only in the livers of transplanted mice, but not in the non-transplanted mice further confirmed the differentiation of fHPCs into functional hepatocytes (Figure 7). In addition, the amplification and detection of human-specific Alu-sequence from the transplanted mouse livers provided molecular evidence for the continued presence of human fetal liver cells in mouse liver at day 80. Sequencing analysis of the amplified Alu gene confirmed the presence of human Alu gene sequence in mouse liver tissues (Supplementary Figure 2). These results suggest that the transplanted human EpCAM-positive DiD-labeled cells continue to survive long-term without eliciting serious unfavorable effects in CLD-SCID mice.
DISCUSSION
This study reports a new application of the lipophilic fluorescent dye DiD for non-invasive in vivo imaging to monitor transplanted fHPCs in SCID mice with CLD. In our earlier clinical study, transplanted fHPCs in end-stage CLD patients were tracked for only 24 h using radio-labeled99mTc-HMPAO post-TX[6]. In our current study, labeling of fHPCs with DiD enabled long-term in vivo monitoring of the transplanted cells, which was further confirmed by ex vivo imaging and immunohistological analysis. Using DiD as a cell label, the transplanted cells could be tracked for 80 d non-invasively in CLD-SCID mouse liver. Our findings revealed that DiD labeling is a relatively simple, safe, and effective approach for long-term and non-invasive tracking of fHPCs post-TX in mouse liver. In addition, our results also showed the efficacy of transplanted human fHPCs as a suitable cell type for improving liver functions similar to earlier studies[11,25].
Long-term in vivo tracking of transplanted cells is currently required to answer several concerns and to reduce existing controversies in cell-based therapies. Now, it has been identified as an essential component of cell-based therapeutic strategies for optimizing the cell number, route of delivery, biodistribution, cell viability post-transplantation, and evaluating the regenerative capabilities, which would aid in accelerating their clinical applications[26]. The depth of penetration, sensitivity, spatial and temporal resolution, ease of availability of the molecular probe, and cost of imaging are some of the important factors to be considered for a suitable imaging approach[26]. Direct labeling of the transplanted cells with fluorescent dyes has been demonstrated in several animal liver disease models to understand their homing and differentiation. For instance, the fluorescent dye PKH26 was used in a CCl4-induced liver injury model in rats to show stem cell migration, proliferation, and expression of liver-specific markers[27,28]. However, these studies did not involve live cell tracking, and only postmortem analysis of the liver tissue sections was performed to identify the presence of labeled cells. Also, due to the potential cross-transfer of the dye to host cells, PKH26 was not considered ideal as a cell tracer, thus limiting its application in transplantation studies[29].
The carbocyanine dyes, CM-DiI, and DiR have been used to label transplanted cells in animal models of liver injury. Although both CM-DiI and DiR proved to be safe for cell tracking applications, monitoring has only been demonstrated in ex vivo settings[30], or if in vivo, the signal was reported to have faded within 5 d after infusion[22]. Hence, the above carbocyanine dyes have not provided sufficient evidence of their long-term applicability for cell tracking in vivo. In contrast, DiD has been demonstrated to be effective for long-term monitoring of labeled neuronal and cancer cells in vitro for up to 4 wk[31,32]. Moreover, DiD at higher concentrations of up to 2 μM also does not affect cell growth, proliferation, migration, and apoptosis, and does not cross-transfer to neighboring cells[33]. In line with this, DiD-labeled MSCs have demonstrated the absence of cytotoxicity and signs of altered functional performance in terms of cytokine production or trilineage differentiation[34], thus assuring the safety of DiD for potential applicability in stem cell tracking. DiD-labeled neural stem cells have also shown promising results following direct injection into the cerebrospinal fluid in vivo[35]. Also, among the Vybrant®dye series (DiR, DiI, CM-DiI, DiD), DiD has demonstrated comparatively high intense fluorescence[32]. Thus, these observations support our choice of DiD for cell labeling and long-term in vivo imaging.
In the present study, DiD did not show any interference with tissue autofluorescence and FITC due to fluorescence emission close to the NIR region. The quick and easy methodology of staining, nontoxicity, no interference with the functionality of the labeled cells, non-diffusion to adjacent cells, and lack of photo-bleaching were identified as the major advantages of DiD labeling. Furthermore, colocalization of CK and c-Met hepatic markers with the DiD-labeled transplanted cells in the recipient mouse livers showed the long-term persistence of transplanted cells. These observations support the involvement of selected markers in supporting the proliferation, survival, and differentiation of transplanted cells in the recipient’s liver regeneration and functional improvements[36-38]. In addition, the detection of hu-ALB-specific expression within the transplanted mouse livers further confirmed that the transplanted fHPCs not only persisted for the longer duration but also differentiated into functional hepatocytes contributing to liver regeneration and functional recovery. In addition, fHPCs successfully attenuated liverfibrosis in mouse liver. However, future studies could aid in deciphering the detailed molecular mechanisms by which fHPCs contribute to liver repair and regeneration.
Overall, the results from our study were supportive of the use of DiD in long-term, non-invasive, in vivo tracking of fHPCs in the recipient’s liver, but there are certain limitations. Although the results indicate the suitability of DiD for monitoring transplanted fHPCs in the liver, quantification of the signal to correlate the cell number or survival was not reported. Future studies using a larger cohort of animals, varying cell numbers, and quantification of the signal intensity in relation to cell doses would be very useful to understand the efficacy and survival. Moreover, optimizing the route of cell delivery, and assessing the dynamic changes in the expression of EpCAM over time will help in addressing the questions on engraftment and differentiation of fHPCs. Lastly, DiD has been proven to be safe and nontoxic with no effect on the metabolic functioning of cells in vitro. However, for regular in vivo imaging applications of DiD, it is essential to evaluate the metabolic cycle of the dye for long-term use. Addressing such issues would make DiD labeling more valuable for wider in vivo cell tracking applications.
CONCLUSION
Monitoring the fate of transplanted cells through in vivo tracking or imaging can help in understanding the homing, engraftment, long-term survival, and function of the transplanted cells. In this preclinical study, DiD-labeled fHPCs showed efficient long-term cell tracking for up to 80 d. The ease of handling, non-toxicity, and long-term signal retention proved to be major advantages in using DiD as a cell labeling agent for non-invasive, long-term tracking of cells both in vivo and ex vivo. These findings could pave the way to unravel the underlying regenerative mechanisms and contribution of exogenously transplanted cells in restoring the structural and functional deficits of the liver in CLD.
ARTICLE HIGHLIGHTS
Research results
This study showed that DiD labeling of human liver cells is easy and efficient for long-term and noninvasive tracking in vivo up to 80 d post-transplantation. Using DiD, the fate of transplanted cells was determined. Transplanted human fetal liver cells were able to provide structural and functional improvement in CLD-SCID mice.
Research conclusions
Monitoring the fate of transplanted cells through DiD-based in vivo live cell imaging can help in understanding the homing, engraftment, long-term survival, and function of the transplanted cells.
Research perspectives
The findings of the current study may pave the way to unravel the underlying regenerative mechanisms and contribution of exogenously transplanted cells in restoring the structural and functional deficits of the liver in CLD.
FOOTNOTES
Author contributions: Khan AA, Pande G, and Tripura C were responsible for the study concept, design, and supervision; Thatipalli AR, Vishwakarma SK, Jose J, and Jerald MK performed the experiments; Tripura C, and Gunda S were responsible for data acquisition and analysis; Tripura C also performed data organization and manuscript writing; Tripura C, Vishwakarma SK, Khan AA, and Pande G performed editing and revision of the manuscript draft.
Supported by Department of Science and Technology (DST), Ministry of Science and Technology, Govt. of India and Indian Council of Medical Research (ICMR), New Delhi, Govt. of India Grants to GP, No. GAP-0220 and No. GAP-0383.
Institutional review board statement: The total fetal liver cells (tFLCs) isolation protocol was approved by the Institutional Ethics Committee (IEC) of Deccan College of Medical Sciences (DCMS), Hyderabad.
Institutional animal care and use committee statement: The study was approved by the Institutional Animal Ethics Committee (Animal trial registration number 20/1999/CPCSEA dated 10/03/1999) of CCMB.
Informed consent statement: Informed consent was obtained prior to sample collection, and cell processing was performed according to the ethical guidelines for the use of human cells.
Conflict-of-interest statement: Dr. Tripura reports grants from the Department of Science and Technology (DST, Ministry of Science and Technology, Govt. of India and Indian Council of Medical Research (ICMR), New Delhi, Govt. of India Grants No. GAP-0220 and No. GAP-0383 to GP during the study period.
Data sharing statement: Data will be shared on request.
ARRIVE guidelines statement: The authors have read the ARRIVE guidelines, and the manuscript was prepared and revised according to the ARRIVE guidelines.
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BYNC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is noncommercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Country/Territory of origin: India
ORCID number: Chaturvedula Tripura 0000-0003-2766-0861; Srinivas Gunda 0000-0003-2053-2330; Sandeep Kumar Vishwakarma 0000-0001-5731-8210; Avinash Raj Thatipalli 0000-0001-6985-3990; Jedy Jose 0000-0003-3155-5386; Aleem Ahmed Khan 0000-0001-7075-9037; Gopal Pande 0000-0002-0730-1389.
S-Editor: Fan JR
L-Editor: Webster JR
P-Editor: Cai YX
 World Journal of Hepatology2022年10期
World Journal of Hepatology2022年10期
- World Journal of Hepatology的其它文章
- Alcohol use disorder and liver injury related to the COVID-19 pandemic
- Hepatic involvement in children with acute bronchiolitis
- Quality of life, depression and anxiety in potential living liver donors for pediatric recipients: A retrospective single center experience
- Immunotherapy for hepatocellular carcinoma: A promising therapeutic option for advanced disease
- Natural history and management or liver aySuncuon ystorage disorders
