Morphological and Molecular Identification of Curvularia geniculata Associated with Leaf Spot Disease of Banana in China
Yanxiang QI, Hong ZHAO, Yixian XIE, Fanyun ZENG, Jun PENG, Xin ZHANG
1. Environment and Plant Protection Institute, Chinese Academy of Tropical Agricultural Sciences/Hainan Provincial Key Laboratory of Pests Detection and Control for Tropical Agriculture, Haikou 571101, China; 2. Center for Research and Development of Fine Chemicals of Guizhou University, Guiyang 550025, China
Abstract [Objectives] The paper was to identify the causal agent of leaf spot of banana in Hainnan, China. [Methods] Fungus isolates were isolated from the diseased tissues, and morphological characteristics of colony and spore were observed. Furthermore, phylogenetic analyses and pathogenicity test were conducted to confirm the pathogen. [Results] The fungus isolates from the diseased tissues was identified as Curvularia geniculata. [Conclusions] C. geniculata is a new pathogen causing leaf spot of banana in Hainan, China. This finding will help to broaden the distribution and host range of C. geniculata, indicating that it poses potential damage to banana in China.
Key words Curvularia geniculata; Leaf spot disease; Musa acuminate; Pathogen identification
1 Introduction
Disease is a main factor limiting banana production. Leaf spot disease is a main banana disease in banana producing areas in China, which mainly damages banana leaves and leads to leaf drying and premature senility of banana plant[1]. A new leaf spot was observed on banana plants at an orchard in Hainan Province, China during 2019 and 2020. The leaf spots occurred sporadically and were mostly confined to the leaf margin, and the symptoms on the leaves were initially reddish brown spots that gradually expanded to ovoid-shaped lesions and eventually became necrotic, dry, and gray with a yellow halo. This study was carried out to identify the causal agent of the leaf spot on banana on the base of colony and spore morphology, phylogenetic analyses and pathogenicity test.
2 Materials and methods
2.1 Fungal isolation and morphological observationTo acquire the pathogen, 30 tissue pieces (15 mm2) of symptomatic leaves from 5 plants were surface-disinfected in 70% ethanol (10 s), and 0.8% NaClO (2 min), rinsed in sterile water 3 times, and transferred to potato dextrose agar (PDA) at 28 ℃. The fungal colonies that developed from the segments were subcultured on PDA. The colony and spore morphology on the media were observed.
2.2 Molecular identification and phylogenetic analysisRe-presentative isolates, morphologically matched the description ofCurvulariageniculata, were grown on PDA at 28 ℃ for 7 d. Genomic DNA was extracted as described by Qietal.[2]. The internal transcribed spacer (ITS) region, large subunit ribosomal DNA (LSU rDNA), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), translation elongation factor 1-alpha (TEF-1α) and RNA polymerase II second largest subunit (RPB2) gene were amplified with primers ITS1/ITS4, LROR/LR5, GPD1/GPD2, EF1-983F/EF1-2218R and 5F2/7cR, respectively[3-8]. The PCR amplification was carried out in a 25 μL reaction mixture containing 0.5 μL of DNA sample, 12.5 μL of PremixTaqTM(TaKaRa Biotech, Dalian, China), 0.5 μL of each primer (20 μM), and 11 μL of nuclease-free sterile distilled water. The PCR conditions were as follows: pre-denaturating at 94 ℃ for 3 min; denaturating at 94 ℃ for 1 min, annealing for 1 min with the corresponding temperatures (55 ℃ for ITS, TEF-1α and RPB2; 53 ℃ for LSU; 56 ℃ for GAPDH), extension at 72 ℃ for 1 min, 35 cycles; final extension at 72 ℃ for 8 min. MEGA7.0[9]was used to construct a maximum likelihood (mL) tree with 1 000 bootstrap replicates, based on a concatenation alignment of five gene sequences of the two isolates in this study as well as sequences of otherCurvulariaspecies obtained from GenBank (Table 1).
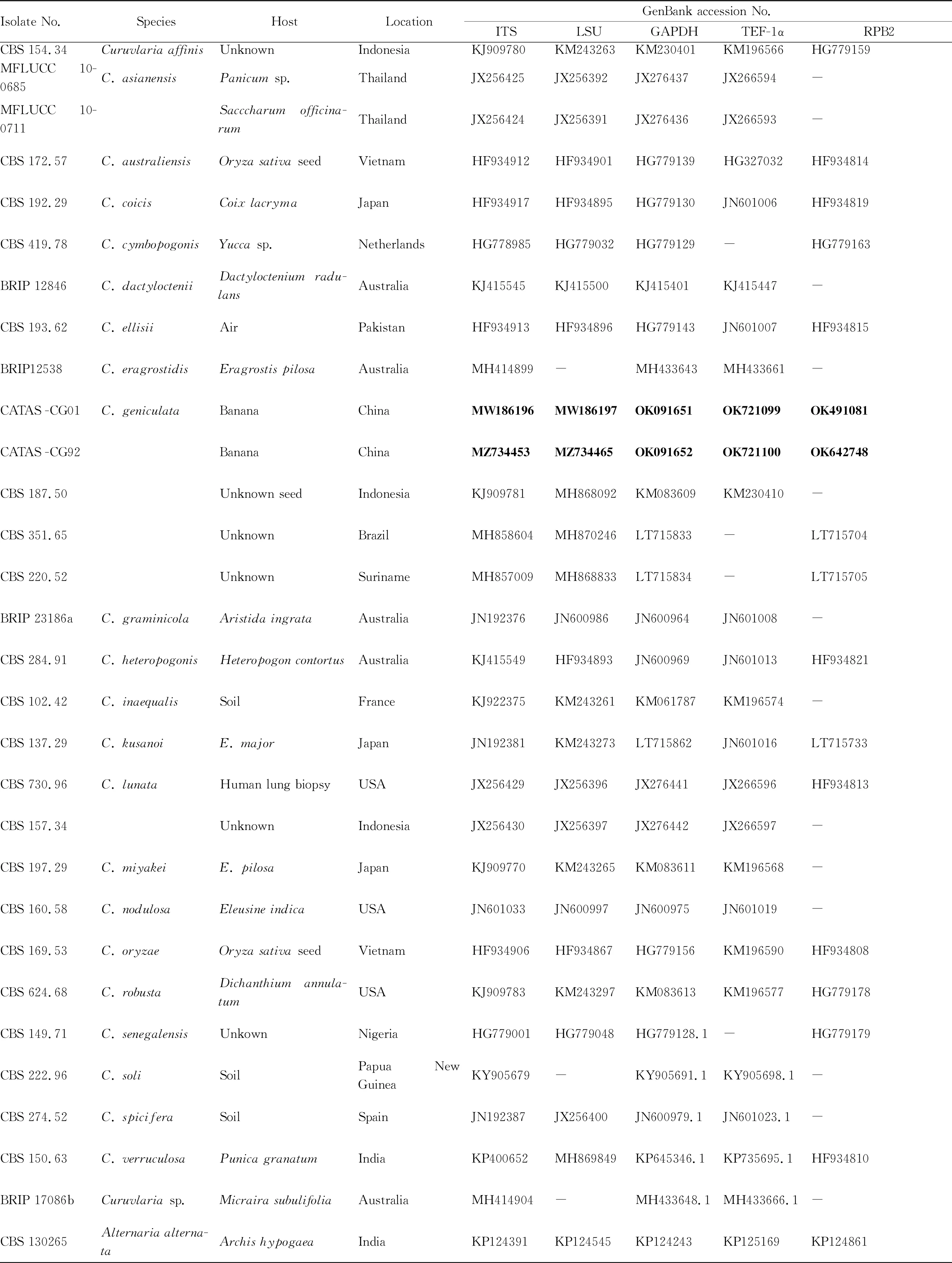
Table 1 Information of isolates used in this study
2.3 Pathogenicity testPathogenicity test was conducted on banana seedlings at the 7-leaf stage. Two leaves from potted plants were stab-inoculated by puncturing into 1 mm using a sterilized needle. Then, 10 μL of conidial suspension (2×106conidia/mL) was inoculated on the surface of wounded leaves, and sterile distilled water was inoculated to equal number of leaves serving as control. Inoculated plants were kept inside a plastic bag for 72 h and then grown in the greenhouse (28 ℃, 90% relative humidity).
The experiment was repeated 3 times. Leaf lesions were examined at 7 d post inoculation, and tissues (15 mm2) were selected for fungal re-isolation from the necrotic lesion edges.
3 Results and analysis
3.1 Fungal isolation and morphological observationFungal colonies appeared and were subcultured after 2 d. Seven days later, subcultures of fungi were grayish green, and then turned fluffy with grey and white aerial mycelium with age (Fig.1). The conidia were brown, erect or curved, fusiform or elliptical, with 3-4 septa with dimensions of (13.75-31.39) × (5.91-13.35) μm (avg. 22.39 μm×8.83 μm). The cells of both ends were small and hyaline, while the middle cells were larger and darker (Fig.1). Morphological characteristics of the conidia matched the description ofC.geniculata(Tracy & Earle) Boedijn[10]. Two representative isolates (CATAS-CG01 and CATAS-CG92) were single-spored and stored in the culture collection of the Environment and Plant Protection Institute, Chinese Academy of Tropical Agricultural Sciences.

Note: A, C. Colony features on PDA; B, D. Pigmentation on reverse plate; E. Conidia on lesions; F. Conidiophores on lesions (Scale bars: E, F=10 μm).
3.2 Molecular identification and phylogenetic analysisTo further confirm the species of the putative pathogen, the ITS region, LSU rDNA, GAPDH, TEF-1α and RPB2 gene of two representative isolates (CATAS-CG01 and CATAS-CG92) were amplified and sequenced with the primers ITS1/ITS4, LROR/LR5, GPD1/GPD2, EF1-983F/EF1-2218R and 5F2/7cR. The sequences were deposited in GenBank (ITS: MW186196 and MZ734453; LSU rDNA: MW186197 and MZ734465; GAPDH: OK091651 and OK091652; TEF1-α: OK721099 and OK721100; RPB2: OK491081 and OK642748). Phylogenetic tree was constructed with MEGA 7.0 based on ITS region, LSU rDNA, GAPDH, EF1-α and RPB2 gene sequences, which confirmed that the representative isolates CATAS-CG01 and CATAS-CG92 wereC.geniculata(Fig.2). Based on the colony, conidia morphology and phylogenetic analyses, the fungus was identified asC.geniculata.
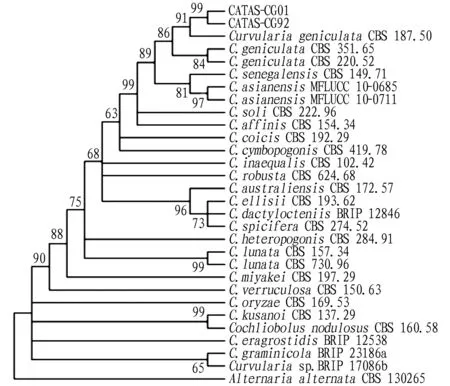
Note: Two representative isolates CATAS-CG01 and CATAS-CG92 were isolated from banana (Musa acuminate) in China by the author and the other reference isolates were from GenBank, which were used as an outgroup. Bootstrap values (1 000 replicates) greater than 50% are shown at the nodes.
3.3 Pathogenicity testThe pathogenicity of this fungus on banana was confirmed by Koch’s postulates. Necrotic lesions on inoculated leaves appeared on inoculated leaves at 7 d post inoculation, whereas the leaves in control remained healthy (Fig.3). Pure cultures ofC.geniculatawere recovered from inoculated leaves, and its taxonomy was confirmed morphologically and molecularly, fulfilling Koch’s postulates.
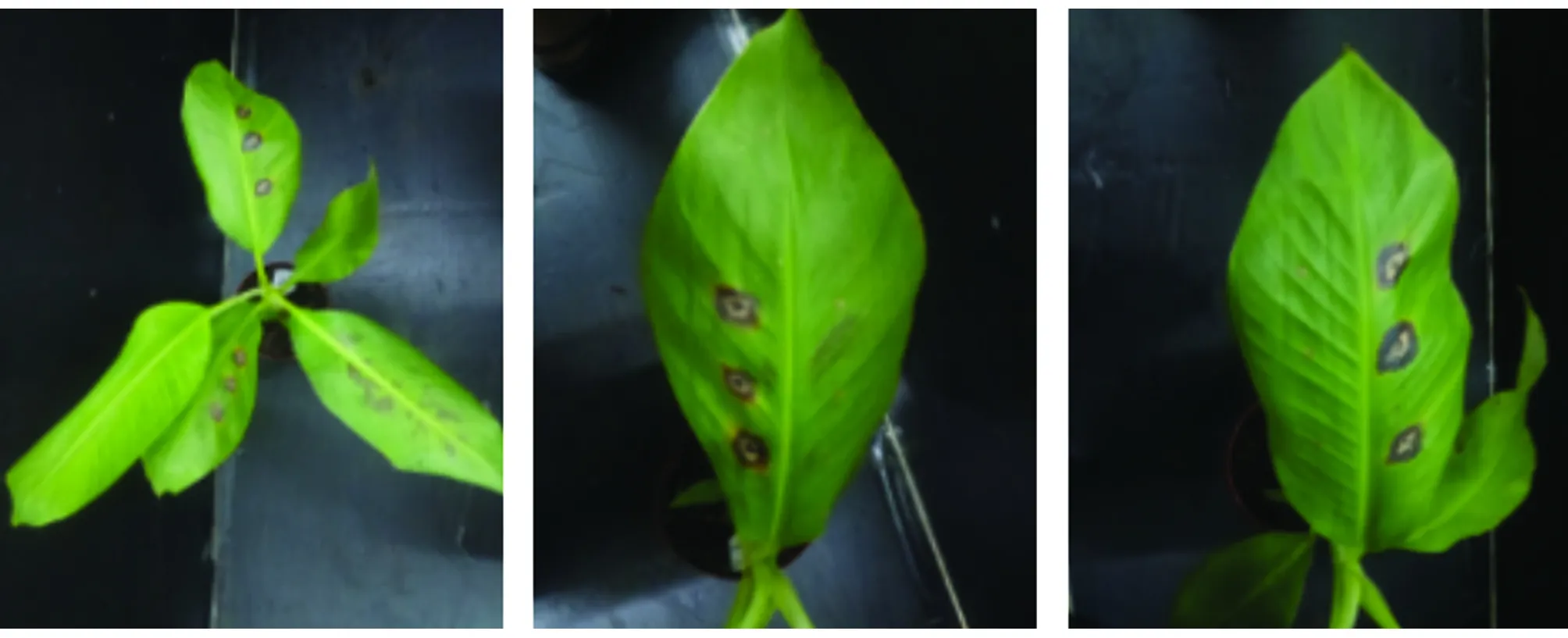
Fig.3 Symptoms in pathogenicity test at 7 d post inoculation
4 Discussion
C.geniculatahas been reported on a wide range of hosts, including banana ‘Lacatan’ in Jamaica[11],Discoreaspp. in Puerto Rico[12],Oryzasativain Indonesia,Trifoliumrepens,ZingbermiogaandAndropogonfurcatusin Japan[10],Semecarpuskathalekanensisin India[13],Eleusineindicain China[14],Microstegiumvimineumin China[15],Paspalumguenoarumin Brazil[16],Saccharumofficinarumin China[17]andZeamaysin China[18], which was observed to be associated with leaf spot or blight. This study provides the first description ofC.geniculataas a pathogen causing
- 植物病虫害研究(英文版)的其它文章
- Effects of Different Herbicides on Weed Control, Agronomic Characters and Grain Quality of Coix lacryma-jobi L.
- Occurrence Dynamics and Countermeasure of Wheat Crown Rot in Weifang City
- Genetic Analysis of Weight per Fruit and Fruit Length in Bitter Gourd
- An Important Insect of the Genus Pseudips: Pseudips mexicanus Hopkins
- Damage Characteristics and Control Techniques of Longhorn Beetles in Coffee Garden
- Syntheses, Structures of A Cobalt (II) Metal-organic Framework Based on 2, 6-di (2’, 4’-dicarboxylphenyl)pyridine and 4, 4’-bis (imidazol-1-ylmethyl)biphenyl-ligands

