Advances in quantitative analysis of astrocytes using machine learning
Demetrio Labate, Cihan Kayasandik
Astrocytes, a subtype of glial cells, are starshaped cells that are involved in the homeostasis and blood flow control of the central nervous system (CNS). They are known to provide structural and functional support to neurons, including the regulation of neuronal activation through extracellular ion concentrations, the regulation of energy dynamics in the brain through the transfer of lactate to neurons, and the modulation of synaptic transmission via the release of neurotransmitters such as glutamate and adenosine triphosphate. In addition, astrocytes play a critical role in neuronal reconstruction after brain injury, including neurogenesis, synaptogenesis, angiogenesis, repair of the blood-brain barrier, and glial scar formation after traumatic brain injury (Zhou et al., 2020). The multifunctional role of astrocytes in the CNS with tasks requiring close contact with their targets is reflected by their morphological complexity, with processes and ramifications occurring over multiple scales where interactions are plastic and can change depending on the physiological conditions. Another major feature of astrocytes is reactive astrogliosis, a process occurring in response to traumatic brain injury, neurological diseases, or infection which involves substantial morphological alterations and is often accompanied by molecular, cytoskeletal, and functional changes that ultimately play a key role in the disease outcome (Schiweck et al., 2018). Because morphological changes in astrocytes correlate so significantly with brain injury and the development of pathologies of the CNS, there is a major interest in methods to reliably detect and accurately quantify such morphological alterations. We review below the recent progress in the quantitative analysis of images of astrocytes. We remark that, while our discussion is focused on astrocytes, the same methods discussed below can be applied to other types of complex glial cells.
Quantitative image analysis:
Automated methods of quantitative image analysis have gained increased recognition in the biomedical field as they allow to objectively extract a multiplicity of features that can be used to interrogate the biomedical data. However, despite the remarkable success in the development of general-purpose algorithms for cell detection and analysis, progress in the automated image analysis of astrocytes has been slow due to the difficulty in adapting such algorithms to images of astrocytes. The challenges in processing microscopy images of astrocytes are due to their broad variability in size and shapes, their unique morphological complexity with processes occurring over multiple scales, and the entangled nature of spatial arrangements they form. During the last few years though, following spectacular advances in machine learning from the applied mathematics and computer science literature, a new generation of more powerful algorithms for biomedical image analysis were introduced, including algorithms specifically targeted to images of astrocytes. InFigure 1A
, we show an image processing pipeline for astrocyte images based on an algorithm recently proposed by Kayasandik et al. (2020). The algorithm is designed to output accurate state-of-the-art astrocyte reconstructions from brain images where astrocytes may be dense; hence, one can compute morphology features of individual cells that can be applied for classification or profiling. Below, we examine the main image processing blocks of the pipeline, with special attention to astrocyte detection and segmentation. In doing that, we also discuss how the analysis methods employed by this algorithm compare with both traditional and more advanced alternative methods. We conclude the paper by discussing future directions and outstanding challenges concerning the automated analysis of astrocytes.Astrocyte detection:
Astrocytes are typically visualized in microscopy images using a glial fibrillary acidic protein marker. Due to their complex morphology, though, conventional methods for cell detection/segmentation based on intensity thresholding perform inconsistently, even when combined with more sophisticated thresholding techniques (Healy et al., 2018). Since these methods are very sensitive to a multiplicity of factors including noise level, image illumination, and cell density, a laborious manual tuning of parameters is often required to guarantee satisfactory performance. By contrast, learningbased methods, especially convolutional neural networks, do provide far superior and more consistent performance. Suleymanova et al. (2018) were the first to propose a deep learning approach to automatically detect astrocytes in microscopy images. This approach, called FindMyCells, is based on an application of DetectNet - a deep learning architecture specifically designed for tasks of object recognition - and was developed using a dataset of 1250 bright-field microscopy images (including over 17,000 cells) that were publicly released in the Broad Bioimage Benchmark Collection. The detection performance of FindMyCells, measured in F1 score, is reported to achieve 0.81 on this dataset, as compared with a conventional method based on automated thresholding where the F1 score is only 0.34. We recall that the F1 score is a measure of binary classification accuracy defined as the harmonic mean of precision and recall; it ranges between 0 and 1, with 1 indicating perfect classification. We remark that the implementation of such a method - as for any learning-based method - requires a training phase, where 1120 out of 1250 annotated images were employed to learn the parameters of the neural network (the remaining 130 images were used for testing). While the detection performance is impressive, one must notice that this result is dependent on the type of training images and detection performance might decrease if the test images are of different types. For instance, it was shown in Kayasandik et al. (2020) that the F1 score of FindMyCells is only 0.41 when computed on a different image dataset consisting of fluorescent images of astrocytes, even though images were acquired at the same magnification level. This drop in performance is due to the difference in image characteristics with respect to the training images, resulting from factors including the different astrocyte populations and the different image modalities (fluorescent microscopyvs
bright field) of the new dataset. To improve detection performance, one should re-train DetectNet using a new training set of images that are of the same type as the images in the new dataset. To generate such a training set, a domain expert has to annotate a sufficient number of images by identifying their spatial location. This observation highlights one of the main limitations of learning-based methods; while they can provide state-of-the-art performance in complex object detection problems, they often require many training samples to achieve competitive performance. This requirement is labor intensive and might be unfeasible if data is scarce due to technical limitations or privacy constraints. On the other hand, more traditional model-based methods do not require training and their results are more interpretable. However, they become more difficult to implement and less performing as the data and the image processing rules they try to impose become more complex.To combine the advantages of the model- and training-based methods, Kayasandik et al. (2020) have proposed a method for astrocyte detection that applies a geometric descriptor called Directional Ratio, whose main idea consists in identifying potential cell bodies associated with pixels of local isotropy in images. Detection is further refined by analyzing the shape characteristics of the astrocyte segmented using a convolutional neural network. The reported F1 score performance is 0.80 on the main image dataset considered in the paper, consisting of fluorescent microscopy images. Remarkably, the F1 score for astrocyte detection on the Broad Bioimage Benchmark Collection dataset used by Suleymanova et al. (2020) is 0.70, showing that this approach is significantly less sensitive to data type than FindMyCells and that performance remains competitive on a rather broad range of images, including an image from bright field microscopy.
Astrocyte segmentation:
Machine learning algorithms currently provide state-of-the-art performance for cell segmentation and deep learning architectures, especially the U-Net architecture and its variants, which have been particularly successful in biomedical applications, as they can be trained with a relatively small number of training images (Falk et al., 2019). We recall that a U-net is a special convolutional neural network where a feature-extracting (encoding) section of the network is followed by a decoding section where pooling operations are replaced by upsampling operators. As a result, the expansive section is approximately symmetric to the contracting section, yielding a U-shaped architecture. For the challenging task of segmenting astrocytes in fluorescent images, it was shown in Kayasandik et al. (2020) that about 100 manually annotated astrocytes were sufficient to train a U-Net architecture to a satisfactory segmentation model.As shown inFigure 1A
, the astrocyte segmentation routine by Kayasandik et al. (2020) is part of an image processing pipeline where astrocytes are first detected and next segmented within an appropriate region of interest. By identifying regions of interest centered on individual cells, this approach also ensures that even cells that are close to each other can be individually detected and segmented. This astrocyte segmentation routine uses a deep learning architecture built using a cascade of two U-nets that were trained using a collection of fluorescent images of astrocytes that were manually segmented by domain experts and were publicly released with the publication. On this dataset, this method achieves an F1 score performance of 0.76, outperforming a basic U-net yielding an F1 score equal to 0.64. As expected, a segmentation routine based on a traditional method such as automated thresholding (Healy et al., 2018) has significantly lower performance and yields a lower F1 score of 0.53.Figure 1B
shows the astrocyte segmentation performance of the method by Kayasandik et al. (2020) as compared to automated thresholding. The figure shows that the first method is also able to separate contiguous cells, while the latter method is unable to capture elongated processes and other complex cellular structures.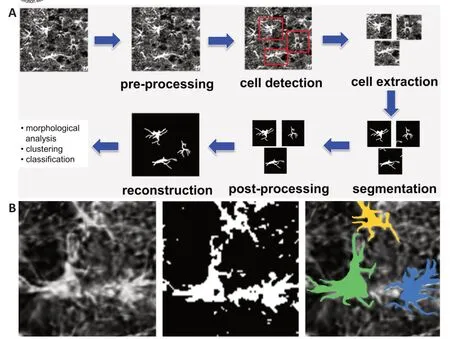
Figure 1|Image processing pipeline for astrocyte images (A) and astrocyte segmentation (B). (A) The pipeline includes a pre-processing stage, followed first by a routine for astrocyte detection and extraction, and next by a routine computing individual astrocyte segmentation. The algorithm outputs an image reconstruction containing all segmented astrocytes; in addition, it provides quantitative information about individual astrocyte morphology and classification. (B) Starting from the left the figure shows: a fluorescent image of astrocytes, the segmentation obtained using the automated local thresholding method of Healy et al. (2018), and the segmentation obtained using the learning-based method of Kayasandik et al. (2020).
Morphological analysis and astrocyte classification:
The ability to automatically detect astrocytes and extract reliable information about their morphology has important practical implications including the development of improved models for the role of astrocytes in the physiology of the CNS and its pathologies. Due to this interest, several automated and semiautomated methods were proposed to analyze the morphology of astrocytes. Kulkarni et al. (2015) and Tavares et al. (2017) in particular, proposed methods to perform morphometric analysis of astrocytes on confocal fluorescence microscopy images of multiplex-labeled (glial fibrillary acidic protein, 4′,6-diamidino-2-phenylindole (DAPI)) brain tissue. In both cases, astrocyte nuclei are identified from the DAPI marker. The algorithms next generate a 3-D arbor reconstruction for each astrocyte either by using a special raytracing algorithm (Kulkarni et al., 2015) or by using a semi-automated pipeline based on the open-source software FIJI-ImageJ and its Neurite Tracer plugin (Taveres et al., 2017). At this point, one can compute standard quantitative arbor measurements, such as the Sholl analysis. More recently, Sethi et al. (2021) introduced a software tool, called SMorph, that integrates multiple functionalities for the quantitative morphological analysis of neurons, astrocytes, and microglia. This method requires that the user manually identifies a region of interest containing a single cell. Next, the software automatically finds the cell body, the main arbors and directly computes several morphometric parameters.A major limitation of the methods described above is that they do not include a capability to automatically detect astrocytes, except in the situation where a DAPI nucleus marker is available. Even in this case though, there is no guarantee that one can reliably associate an astrocyte to a DAPI marker. Furthermore, the astrocyte reconstruction generated by these methods typically captures only the main cell processes (missing features that have a potentially significant physiological role) and there is no systematic validation of the reconstruction process. By contrast, one of the advantages of the new generation of algorithms such as that image processing pipeline inFigure
1A
is that it outputs individually segmented astrocytes starting from an image containing multiple cells, without the need of a nucleus marker, even when astrocytes are densely packed. Working on individually segmented astrocytes, it is then straightforward to compute any multiple morphometric features and possibly to integrate the astrocyte segmentation algorithm into a highcontent image screening pipeline. An additional advantage of this learning-based approach is the possibility of extracting morphological features via representation learning. Namely, since a deep neural network is used for segmentation, shape features can be associated with the hidden layer coefficients of the network. This type of approach has already been explored for other biomedical applications and offers a promising avenue for an unbiased approach to the morphological analysis of astrocytes.Conclusion:
As compared with other areas of biomedical image analysis, such as neuronal imaging, the development of automated methods targeted at the analysis of images of astrocytes is still in its infancy. In the opinion of the authors, a major outstanding obstacle to the development of the field is the lack of publicly available image databases to be used for benchmarking methods of astrocyte detection and segmentation - currently, the only publicly available databases of astrocyte images are the Broad Bioimage Benchmark Collection set published by Suleymanova et al. (2018) and the set released by Kayasandik et al. (2020), where the first one is only annotated for detection and not for segmentation. By contrast, the field of neuronal imaging has benefited enormously from major projects such as NeuroMorpho (Ascoli et al., 2007) that provide public-access archives of neuronal images and digital reconstructions of neuronal morphology. It is highly desirable that a similar effort would be carried out for astrocytes and possibly other glial cells, as this would have a dramatic and transformative impact in the field.This work was supported by National Science Foundation grants Division of Mathmatical Science 1720487 and 1720452 (to DL).
Demetrio Labate
, Cihan Kayasandik
Department of Mathematics, University of Houston, Houston, TX, USA (Labate D)Department of Computer Istanbul Medipol University, Istanbul, Turkey (Kayasandik C)
Correspondence to:
Demetrio Labate, PhD, dlabate2@Central.UH.EDU.https://orcid.org/0000-0002-9718-789X(Demetrio Labate)
Date of submission:
January 13, 2022Date of decision:
March 9, 2022Date of acceptance:
March 21, 2022Date of web publication:
June 2, 2022https://doi.org/10.4103/1673-5374.346474
How to cite this article:
Labate D, Kayasandik C (2023) Advances in quantitative analysis of astrocytes using machine learning. Neural Regen Res 18(2):313-314.
Availability of data and materials:
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Open access statement:
This is an open access journal, and articles are distributed under the terms of the Creative Commons AttributionNonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
Open peer reviewer:
Xu-Feng Yao, Shanghai University of Medicine and Health Sciences, China.
Additional file:
Open peer review report 1.
- 中国神经再生研究(英文版)的其它文章
- Kainate receptors in the CA2 region of the hippocampus
- Nicotinic acetylcholine signaling is required for motor learning but not for rehabilitation from spinal cord injury
- Understanding the timing of brain injury in fetal growth restriction: lessons from a model of spontaneous growth restriction in piglets
- MicroRNA-based targeting of the Rho/ROCK pathway in therapeutic strategies after spinal cord injury
- Intrathecal liproxstatin-1 delivery inhibits ferroptosis and attenuates mechanical and thermal hypersensitivities in rats with complete Freund’s adjuvant-induced inflammatory pain
- Commentary on “PANoptosis-like cell death in ischemia/reperfusion injury of retinal neurons”

