Synthesis, Crystal Structure, DNA Binding Ability of a New Bis-Pyridine Pendant-Arms Mn(Ⅱ) Complex
, , , ,
(1.School of Chemical and Environmental Engineering, Wuhan Polytechnic University, Wuhan 430023, China;2.Sichuan Institute of Product Quality Supervision and Inspection, Chengdu 610100, China)
Abstract:A new dinuclear manganese(Ⅱ) complex with bis-pyridine pendant-arms was synthesized by condensation between 3,3′-((ethane-1,2-diylbis((pyridin-2-ylmethyl)azanediyl)) bis(methylene))bis(2-hydroxy-5-methylbenzaldehyde) and 2-hydroxy-1,3-diaminopropane in the presence of manganese(Ⅱ). The complex was determined by X-ray diffraction single crystal structure analysis, and the corresponding formula is [Mn2(C37H43N6O6)]·(ClO4)2. The results indicate that the complex crystallizes in monoclinic, space group P21/c, with a=1.096 50(19) nm, b=1.419 5(3) nm, c=3.109 4(5) nm, β=108.153(5)°. The crystal structure shows that the two manganese(Ⅱ) ions in the phenolbased macrocyclic dinuclear complex are coordinated with (Namine)2(Nimine)2O3 and (Nimine)2O4 sites, the corresponding geometry around each manganese(Ⅱ) center are decahedron and distorted octahedron. Two manganese(Ⅱ) centers are equivalently bridged by the phenolic oxygens and an acetate radical with the intermetallic separation of 0.331 6 nm. Moreover, the binding ability of the complex toward calf thymus DNA were analyzed by voltammetric and viscosity method, which indicate the bonding mode between them is weak intercalation.
Key words:dinuclear manganese(Ⅱ) complex; macrocyclic metal complex; crystal structure; DNA binding ability
0 Introduction
The macrocyclic metal complexes have been given much attention for their special bioregulatory activities[1-2]. In nature, they are widely found in organisms, such as hemoglobin porphyrin iron derivatives, chlorophyll porphyrin magnesium derivatives, and many kinds of metalloenzymes[3]. Since the early 20th century, the design and synthesis of metallacycles with one or more metal cations have been developed enormously[4-6]. The macrocycles coordinate with the transition metal forming stable complexes by relatively weak forces such as hydrogen bonds, ion-dipole and dipole-dipole interactions, van der Waals interactions, etc[7]. Their positive application in the fields of catalysts, biology and photochemistry attracted the interests of researchers[8]. A number of polynuclear Schiff base functionalized pendant arms metal systems have been structurally characterized in relation to bioinorganic models, and their bioactivity to calf thymus(CT) DNA, pBR322 DNA and 4-nitrophenyl phosphate disodium salt hexahydrate (pNPP) have also been investigated[9]. In the paper, a new dinuclear manganese(Ⅱ) complex with bis-pyridine pendant-arms has been prepared in medium yield. The crystal structure shows that this 19-memebered metallacycle derived from aromatic meta-dialdehydes and containing 104 atoms including 14 oxygen atoms. DNA binding ability have been investigated on the basis of voltammetric and viscosity method, the researches can obtain the interaction between DNA and drug molecules, which are of the frontier fields in modern bioactivity research[10]. Furthermore, these binding studies have been very useful in designing new and promising drug for clinical use and developing sensitive chemical probes of nucleic acid structure[11].
1 Experimental section
1.1 Synthesis of 3,3-((ethane-1,2-diylbis((pyridin-2-ylmethyl)azanediyl))bis(methylene))bis(2- hydroxy-5-methylbenzaldehyde) (H2L)
N1,N2-bis(pyridin-2-ylmethyl)ethane-1,2-diamine and 3-(bromomethyl)-2- hydroxy-5-methyl were prepared according to the literature methods[12-13].
To a solution of 3-(bromomethyl)-2-hydroxy-5-methylbenzaldehyde (5.2 g, 22 mmol) and Et3N(4.2 g, 40 mmol) in dry CH2Cl2(50 mL), a solution of N1,N2-bis(pyridin-2-ylmethyl) ethane-1,2-diamine (2.5 g, 10 mmol) in dry CH2Cl2(40 mL) was added dropwise. The red solution was stirred for 18 h at room temperature. The solution was washed with water, then, the solvent was removed from the organic phase under reduced pressure. HCl (5 mol/L, 50 mL) was added to the solution and extracted with CH2Cl2. The aqueous phase was adjusted to pH=7 with aqueous NaOH (8 mol/L). The resultant mixture was extracted with CH2Cl2again. Removal of the CH2Cl2to afford a brown oil which was purified by column chromatography upon silica gel. Yield: 3.2 g.1HNMR (300 MHz, CDCl3) δ 11.09 (br s, 2H), 10.19 (s, 2H), 8.54 (d,J=4.6 Hz, 2H), 7.61 (d,J=1.1 Hz, 2H), 7.37 (s, 2H), 7.33~7.07 (m, 6H), 3.74 (s, 4H), 3.66 (s, 4H), 2.75 (s, 4H), 2.23 (s, 6H). NMR (75 MHz, CDCl3) d 192.25 (s), 158.64 (s), 157.48 (s), 148.54 (s), 137.18 (s), 136.43 (s), 128.44 (s), 127.74 (s), 124.63 (s), 122.84 (s), 121.92 (d, J=13.3 Hz), 58.97 (s), 54.57 (s), 50.12 (s), 19.93 (s).
1.2 Synthesis of the complex
To a H2L (0.135 g, 0.25 mmol) ethanol solution, Mn(OAc)2·4H2O(0.047 g, 0.25 mmol) in ethanol (10 mL) was added dropwise. The mixture was stirred for 2 h, a solution of 1,3-diamino-2-propanol (0.023 g, 0.25 mmol) in ethanol (10 mL) was added slowly, stirred for another 2 h. The suspension turned a brown clear solution. Then a solution of Mn(ClO4)2·6H2O (0.091 g, 0.25 mmol) was added to above solution and stirred for 8 h. After filtration, the green precipitate was collected and dissolved in the solution of acetonitrile and ethanol (v∶v=2∶1), brown block crystals suitable for the X-ray measurement were obtained by evaporation of the filtrate at room temperature for 14 d. Yield: 0.148 g (65%). Anal. Calc. for C37H43Cl2Mn2N6O14(%):C, 48.67; H, 5.19; N, 9.20. Found: C, 48.71; H, 5.02; N, 9.36.
The crystal structure of the complex was carried out on a Bruker AXS SMART diffractometer using graphite monochromatized Mo-Kα. Data reduction and cell refinement were reported by SMART and SAINT Programs[14]. The structure was solved by direct methods (Bruker SHELXTL) and refined onF2by the full-matrix least-squares method[15]. Hydrogen atoms were located geometrically and refined in riding mode. The non-H atoms were refined with anisotropic displacement parameters. Calculations were performed using the SHELX-97 crystallographic software package.
2 Results and discussion
2.1 IR spectra
2.2 Crystal structure
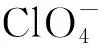
Similarity to the reported heterobinuclear Cu(Ⅱ)Ni(Ⅱ) complex with benzyl pendant-arms, the two pyridine pendant-arms of the titled complex is situated in the same sidepiece of the mean molecular plane[9]. The two Mn(Ⅱ) ions with (Namine)2(Nimine)2O3and (Nimine)2O4metal-binding sites own different coordination environments due to the diverse size of cavities provided by the asymmetry structure of ligand[8]. Mn1 is bound by the O and N atoms belonging to ligand(O5, O6, N1, N2, N3, N4) and acetate(O8) to form a seven-coordinate complex with two six-membered chelate rings (ring 1: Mn1-O6-C16-C17-C15-N3; ring 2: Mn1-N4-C34-C33-C35-O5) and three five-membered chelate rings(ring 1: Mn1-N1-C5-C6-N3; ring 2: Mn1-N3-C7-C8- N4; ring 3: Mn1-N4-C9-C10-N2). The coordination geometry of Mn1 can be described as a decahedron. Mn2 is bound by the O and N atoms belonging to ligand (O5, O6, N5, N6), water(O16) and acetate(O7) to form a six-coordinate complex with three six-membered chelate rings (ring 1: Mn2-O6-C16-C22-C23-N6; ring 2: Mn2-N5-C26-C25-C24-N6; ring 3: Mn2-O5-C35-C28-C27-N5). Mn2 in the six-membered chelate rings deviate from the planes for a mean value of 0.000 12 nm. It adopts a distorted octahedron geometry with two imine and two phenoxide donors lying on the equatorial plane, the corresponding bond angles are in the range from 0.187 1(4) nm to 0.201 7(6) nm. The axial positions are occupied by a water molecule and a acetate group, which weakly bond to Mn2[18].

FormulaC37H43Cl2Mn2N6O14Formula weight976.55Crystal systemMonoclinicSpace groupP21/ca/nm1.096 50(19)b/nm1.419 5(3)c/nm3.109 4(5)β/(°)108.153(5)Volume/nm34.598 8(15)Z4D(calc)/(g·cm-3)1.410Mu (Mo Kα)/mm0.732 F(000)2 012.0Crystal size/mm0.20×0.22×0.24Temperature/K293Mo Kα radiation/nm0.071 073θ range/(°)1.1, 26.0Nref, Npar8 036, 590Tot., uniq. data R(int)23 208, 8 036, 0.07Observed data [I>2.0 σ(I)]3 253R, wR2, S0.063 6, 0.187 8, 0.966
Two Mn(Ⅱ) centers are unequivalently bridged by the phenolic oxygens and an acetate radical with the intermetallic separation of 0.331 6 nm. The dihedral angle between the (Namine)2O2and (Nimine)2O2planes is 8.857(117)°. The two pyridine pendant-arms linked to Mn1 nearly perpendicular to each other with Mn—N distances of 0.243 8(5) nm (Mn1—N1) and 0.224 9(6) nm (Mn1—N2), respectively. The distances between Mn(Ⅱ) center and nitrogen atoms are longer than that of mononuclear Co(Ⅱ) complex[19].
2.3 DNA binding studies
The cyclic voltammetric behaviors of the complex at different scan rates in the sweep range from -1.4 V to 0 V are shown in Fig.3. A pair of redox peak was found at every experimental scan rate and considered as Mn(Ⅱ)Mn(Ⅱ)/Mn(Ⅱ)Mn(Ⅲ) redox couple. For the scan rate of 100 mV·s-1, the anodic peak potential (Epa) and the cathodic peak potential (Epc) are -0.636 V and -0.826 V, respectively. Half-wave potentials (E1/2), taken as the average ofEpaandEpc, is -0.731 V, which is 0.030 V lower than that ofE1/2in the analogous heterodinuclear Zn(Ⅱ)NiL(Ⅱ) complex[20].The difference can be assigned to the diverse coordination environment around metal ions. The separation of the anodic and cathodic peak potentials(ΔEp) is greater than the Nernstian values of 59 mV for one electron redox system. The nonequivalent current intensity of cathodic and anodic peaks indicate that it’s a quasi reversible electrochemical process[21]. The very slight variations ofipc/1/2values at the scan rates from 50 mV·s-1to 200 mV·s-1indicates that this process is mainly diffusion controlled[22].
Upon the addition of CT-DNA, it exhibits one metal center oxidation peak at -0.591 V of Mn2+/Mn3+and two successive ligand-based reduction (pyridine0/pyridine-1and pyridine-1/pyridine-2) at -0.789 V and -1.082 V, indicate that the oxidation potential based on metal center is stronger than that of pyridine arms in anode, and the reduction potential based on metal center is weaker than that of pyridine arms in cathode[23]. Stronger electron-donor capacity of the pyridine arms in complex renders the metal center reduction is difficult in the DNA-binding process.
The mode of complex-DNA interaction can be judged from the variation of formal potential. In general, the anodic shift in formal potential is caused by the intercalation of the complex into the double helical structure of DNA, and the cathodic shift in formal potential is the reason of electrostatic interaction between complex and phosphate of DNA backbone[24]. In the condition,Epawas shifted slightly by 0.06 V toward anode andIpawas dropped by 70.2%, indicating the weak intercalation. Similar benzyl or thiophenoethyl pendant-arms complexes have higher binding activity than the present pyridine pendant-arms complexes[25-26].
Viscosity method is also an effective tool in clarifying the mode of interaction of complex with DNA[27]. Experiments were carried out using a capillary viscometer at (25.0±0.1) ℃. Each set of data measured in triplicate, averages are presented as (η/η0)1/3versus molar ratio of complex to DNA, whereηis the viscosity of CT-DNA in the presence of the complex andη0is the viscosity of CT-DNA alone. As shown in Fig.4, the specific viscosity of the DNA increases slightly with the increasing amounts of complex. This again suggests that the binding mode between the complex and DNA should be a weak intercalation mode[9].
3 Conclusion
A new asymmetrical bis-phenol pendant-armed macrocyclic binuclear Mn(Ⅱ) complex by template-directed synthesis method was successful synthesized in this paper. The obtained complex was characterized by IR spectrum, elemental analysis, and X-ray diffractions single crystal structure analysis. The crystal structure shows that the two manganese(Ⅱ) ions are coordinated with (Namine)2(Nimine)2O3and (Nimine)2O4sites, respectively. Two metal centers are equivalently bridged by the phenolic oxygens and an acetate radical with the intermetallic separation of 0.331 6 nm. The investigations of electrochemical and viscosity experiments indicate that this complex interacts with DNA by a weak intercalation mode compared with similar benzyl or thiophenoethyl pendant arms complexes, which is a strong evidence of the fact that the functionalized pendant arms can significant impact the binding ability.

