Antagonistic Effects of Sphingomonas and Pseudomonas aeruginosa on 4 Kinds of Pathogenic Bacteria of Ginseng
Hairu YU, Feifan YAN, Yunlong WANG, Xinying TONG, Di CHEN, Qiang YE, Renzhe PIAO, Hongyan ZHAO*
1. Yanbian Characteristic Industry Development Center, Yanji 133002, China; 2. College of Agriculture, Yanbian University, Yanji 133002, China; 3. Longjing Agricultural Technology Extension Station, Longjing 133400, China; 4. Yanbian Korean Autonomous Prefecture Academy of Agricultural Sciences, Longjing 133400, China
Abstract [Objectives] To explore effective biocontrol methods for diseases in the process of ginseng cultivation, and develop an efficient and environmentally friendly biocontrol agent. [Methods] In this study, 2 strains were isolated from biogas slurry, and Cylindrocarpon destructans (XF), Fusarium solani (GF), Botrytis cinerea Pers (HM) and Alternaria panax Whetz (HB) were used as test materials. The strains were isolated and identified by dilution plate method, 16S rDNA sequence identification method, confrontation culture method, filter paper method and ultraviolet spectrophotometer method, and the bacteriostatic activity and bacteriostatic rate were tested. [Results] Strain 15 (Sphingomonas) and strain 19 (Pseudomonas aeruginosa) were screened out through identification and analysis, and they grew stably within 8-10 d. The bacteriostatic rates of strain 15 against A. panax and B. cinerea were 47.37% and 43.40%, respectively, and the bacteriostatic rates of strain 19 against A. panax and B. cinerea were 62.30% and 63.27%, respectively. The bacteriostatic activity of the extract of strain 19 increased with the increase of OD600 value, and the bacteriostatic effect was optimal when the OD600 value was in the range of 0.8-1.0, up to 70%, so it had a strong biocontrol potential. [Conclusions] This experiment provides convenience for more effective inoculation, establishes a fast, simple and accurate method for the determination of the best bacteriostatic rate of P. aeruginosa culture solution to HM, and lays a foundation for large-scale culture of P. aeruginosa culture solution. Besides, it is expected to provide a theoretical basis for the efficient control of ginseng B. cinerea in field production, use it for the prevention and control of ginseng shoot diseases, and provide a reference for the efficient and diverse development of biocontrol agents for ginseng shoot diseases.
Key words Panax ginseng C. A. Meyer, Ginseng diseases, Antagonistic bacteria, Screening, Identification
1 Introduction
Ginseng (PanaxginsengC. A. Meyer) is a medicinal plant belonging to the genusPanaxin the family Araliaceae. It is a traditional precious Chinese herbal medicine and reputed as the king of medicinal herb[1-2]. In China, ginseng cultivation is mainly concentrated in the three northeastern provinces. Ginseng black spot and grey mould are the main diseases of ginseng shoots. They become more and more common and serious, not only influencing the ginseng yield but also reducing the ginseng quality, as well as the healthy and green development of the ginseng industry[3-4]. Ginseng diseases caused by pathogenic microorganisms are becoming more and more serious. At this stage, the control measures of plant diseases mainly rely on chemical control[5]. However, the long-term and large-scale application of pesticides has become increasingly prominent, such as chemical residues, environmental pollution, and emergence of drug-resistant pathogenic strains. In recent years, with the increasing attention to the ecological environment, biocontrol has been widely used in the prevention and control of ginseng diseases due to its benefits of safety, economy, high efficiency, and environmental protection[6-8]. Studies have shown thatTrichodemaspp.,Bacillus,Pseudomonas,etc., can control plant diseases through nutrient competition, space competition, parasitism, and synthetic antibiotics[9-11]. Among them, the biocontrol ofTrichodemaspp. andBacillushas been widely used. Because of wide distribution,HalomonasandPseudomonasare easy to apply. Many of strains can effectively inhibit a variety of pathogenic microorganisms and have become valuable biocontrol bacteria and rhizosphere growth-promoting bacteria, but there are few reports about the application ofHalomonasandPseudomonasin the biocontrol of ginseng diseases[12-13]. In this study, we isolated Strain 15 (Sphingomonas) and strain 19 (Pseudomonasaeruginosa) from biogas slurry. We identified their bacteriostatic effect and varieties. We used the confrontation culture method to quickly verify the bacteriostatic activity of the strains, and used the indoor confrontation culture method, the filter paper method and the spectrophotometer method to further verify the bacteriostatic activity of the strain 19 against ginsengBotrytiscinereaPers (HM). It is expected to provide convenience for more effective inoculation, to establish a fast, simple and accurate method for determining the best bacteriostatic rate ofP.aeruginosaculture solution against HM, lay a foundation for large-scale culture ofP.aeruginosaculture solution, provide a reference for the development of ginseng disease biocontrol agents, and to provide a reference for the green prevention and control of ginseng diseases.
2 Materials and methods
2.1 Test bacteriaGinsengCylindrocarpondestructans(XF),Fusariumsolani(GF),BotrytiscinereaPers (HM) andAlternariapanaxWhetz (HB) were provided by pathology laboratory of College of Agriculture, Yanbian University.
2.2 Culture medium(i) Preparation materials of PDA medium: 200 g of potato, 15 g of glucose, 18 g of agar, 1 000 mL of distilled water, and natural pH.
(ii) Preparation method of PDA medium: weighed 200 g of potatoes, washed, peeled and cut into pieces, added 1 000 mL of water to boil for 30 min, filtered with gauze, added 15 g of glucose and 18 g of agar, fully dissolved and filtered with 8 layers of gauze while hot, separated placed and sterilized under high pressure steam at 115 ℃ for 15 min.
(iii) PCS solid medium: 5.0 g of peptone, 5.0 g of cellulose, 5.0 g of NaCl, 2.0 g of CaCO3, 1.0 g of yeast powder, 18 g of agar, 1 L of distilled water, and pH 7.2-7.4.
(iv) LB liquid medium: 10 g of tryptone, 5 g of yeast extract, 10 g of sodium chloride, 1 000 mL of distilled water, and pH 7.0.
2.3 Screening, isolation and purification of target strains
Took 10 mL of the compound bacterial solution, put it into a sterilized 150-mL conical flask, added 90 mL of sterilized physiological saline to make a mixed solution with a dilution of 10-1, shook up and mixed well. Took 1 mL of the mixture and added 9 mL of sterilized physiological saline to make a dilution of 10-2. In this way, prepared the mixture of dilution of 10-3, 10-4, 10-5, and 10-6, then spread 0.1 mL of each on PCS solid medium, and incubated at 37 ℃ for 4 d at an inverted constant temperature incubator, inoculated the grown colonies on PCS medium again, and conducted purified incubation by plate streaking method.
2.4 Identification of strain speciesAfter morphological observation and screening of the isolated strains, we screened them again by Gram staining and microscope observation, and selected Gram-negative bacteria and sent them to Shanghai Sangon Biotech Co., Ltd. for sequencing analysis. PCR primer sequences: 27 F, 1492 R. We compared and analyzed the sequencing results using the BLAST database, and selected the most similar strain as the reference strain. With the aid of MEGA 7 software, we constructed a phylogenetic tree and plotted a 16S rRNA phylogenetic tree.
2.5 Culture of strain 19 seed liquidWe spread the activated strain 19 on a plate for expansion. After 48 h, we washed the bacteria on the plate with 200 mL of LB, placed at 24 ℃, 200 r/min constant temperature shaking for 24 h, and prepared bacterial suspensions with different concentration gradients. Then, we measured the absorbance (OD) of bacterial solution using a spectrophotometer (λ=600 nm). Took the bacterial solution with anOD600of 0.1 in a conical flask. After 6 h, measured theOD600every 2 h (24 h in total). The above operations were carried out under sterile conditions.
2.6 Bacteriostatic activity of strainsPlate confrontation culture method: selected bacterial strains to activate on PCS medium for 3-5 d. Ginseng XF, GF, HM and HB pathogens were activated on PDA medium for 8 d. Used a hole puncher to punch out 6 mm petri dish on the edge of each pathogen and put them in the center of a new PDA plate. The activated antagonists (2 symmetrical points) were inoculated to the 1 cm on both sides of the 4 kinds of ginseng petri dishes, respectively. Each treatment was repeated 3 times, with no antagonistic bacteria as the control (CK), cultured at 20 ℃ constant temperature for 13 d, and the diameter and width of the pathogenic bacteria colonies using the formula (1) were measured every day from the third day to calculate the bacteriostatic rate using the formula (2).
Filter paper method[14]: selected strain 19 seed liquid to culture the bacterial solutions with differentOD600values at the 0, 6, 10, 14, 22 and 24 h respectively. Dipped the sterilized filter paper with a diameter of 6 mm into 0.02 mL of each bacterial solution, and placed on the PDA medium that has been inserted with pathogenic bacteria by 2 points per dish. Placed a piece of filter paper symmetrically at 1 cm on both sides of the petri dish. Each treatment was repeated 3 times, with no antagonistic bacteria as the control (CK), cultured at 20 ℃ constant temperature, and the diameter width of the pathogenic bacteria colonies using the formula (1) were measured every day from the third day to calculate the bacteriostatic rate using the formula (2).
Colony growth width=Average width of 3 times-6 mm
(1)
Bacteriostatic rate (%)=[(Control colony growth width-Treatment colony growth width)]/Control colony growth width×100
(2)
3 Results and analysis
3.1 Species identification of strain 15 and strain 19We carried out Gram staining and microscope observation on the isolated strains, and performed BLAST analysis and comparison of the 16S rRNA genes of strains 15 and 19 in the NCBI database. From the analysis of the phylogenetic tree (Fig.1), it can be seen that the 16S rRNA sequence of strain 15 had a higher homology toSphingomonas, so we preliminarily identified that strain 15 wasSphingomonas; the 16S rRNA sequence of strain 19 had a higher homology toPseudomonas, so we preliminarily identified that strain 19 wasPseudomonas.
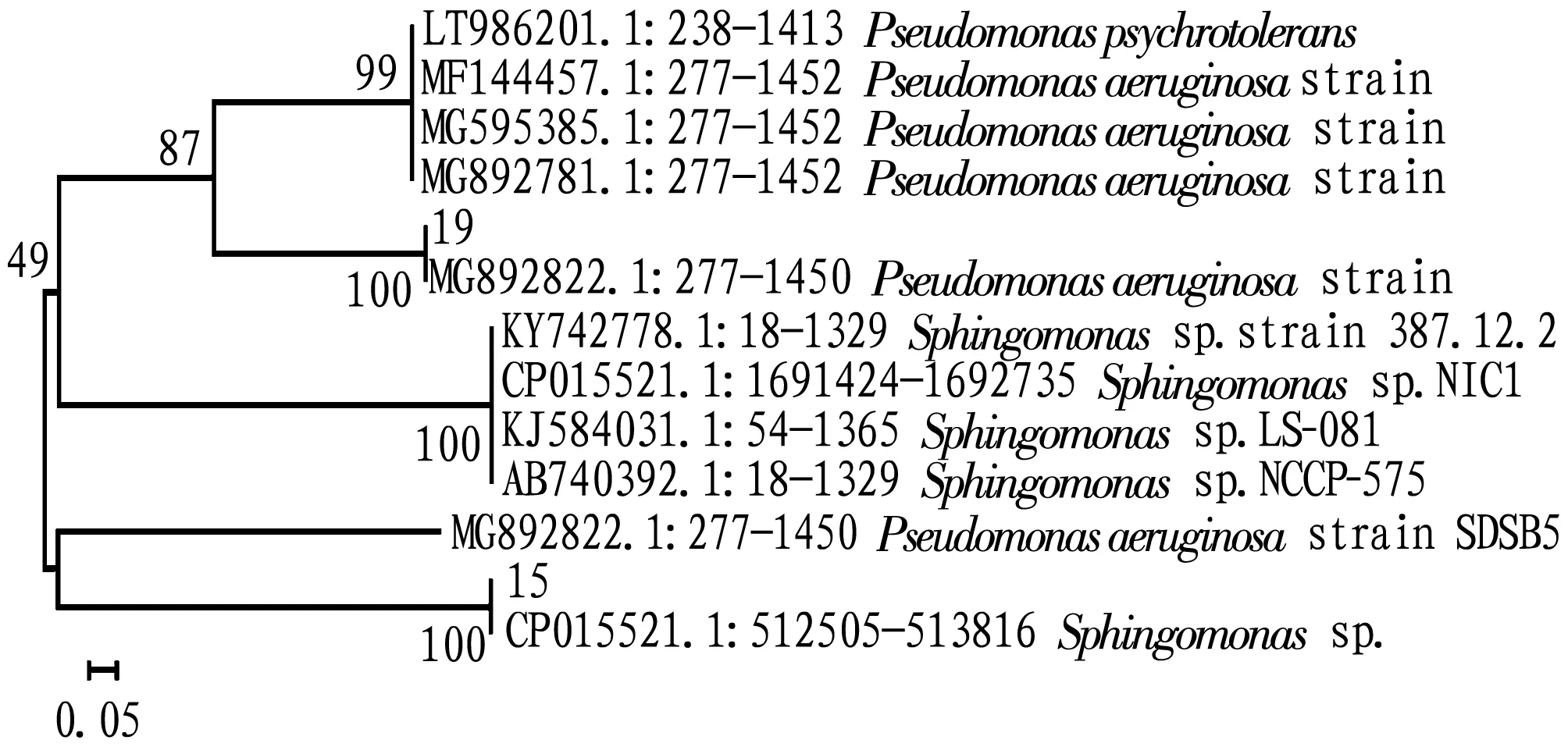
Fig.1 Phylogenetic tree constructed based on the 16S rRNA sequences of strains 15 and 19
3.2 Growth of pathogenic bacteria colonies in different days of treatment strains
3.2.1Colony growth of 4 kinds of ginseng pathogens by the same treatment.
As shown in Fig.2, the growth of colony width in the control group was GF>HB>XF>HM; the colony width growth of treatment strain 15 and treatment strain 19 was GF>XF>HB>HM. On the whole, GF colonies grew the fastest and HM colonies grew the slowest. For treatment strain 15 and treatment strain 19, the growth was relatively stable in 8-10 d, and the HB treated with strain 15 and the HM treated with strain 19 had an obvious increasing trend in 10-11 d.
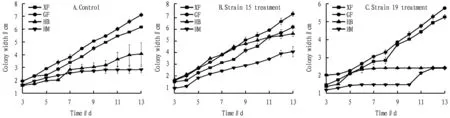
Fig.2 Variation trend of colony width in 3-13 d
3.2.2Effects of different treatments on the colony growth of the same ginseng pathogen. As shown in Fig.3, both strain 15 and strain 19 had no obvious inhibitory effect on XF and FP, but had visible inhibitory effect on HB and HM. In particular, strain 19 had significant inhibitory effect on HB and HM, and the growth was relatively stable in 8-10 d. Besides, in 10-12 d, strain 15 had a significant increase in the colony width of HB, and strain 19 had a significant increase in the colony width of HM.
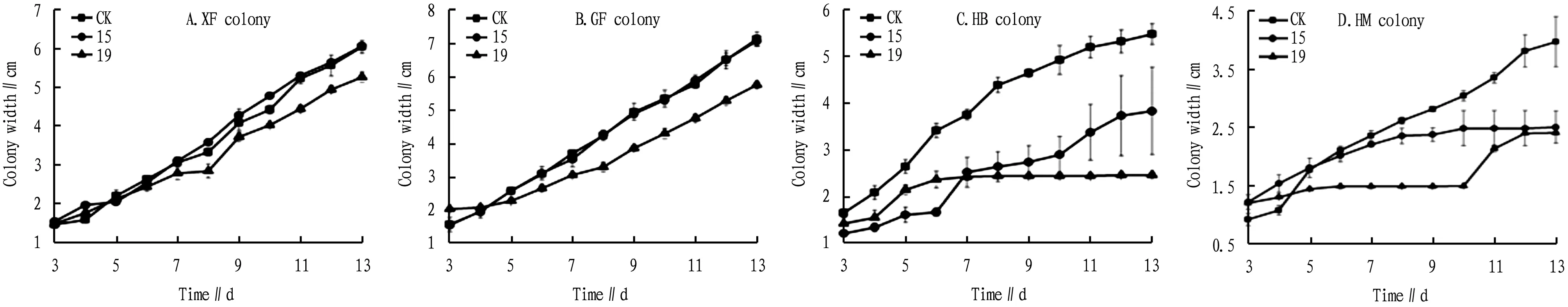
Fig.3 Variation trend of different colony widths in the three treatments in 3-13 d
3.3 Variation in the bacteriostatic activity of the two strains in 3-13 dAs shown in Fig.4, the bacteriostatic rate tended to be stable in 8-10 d, which is consistent with the results in Section3.2. Strain 15 had the highest bacteriostatic rate of XF on the 5thd, but most of them were negative, which was not universal; the bacteriostatic rate of strain 15 against GF changed very little, and the research was of little significance; on the 6thd, the bacteriostatic rate of strain 15 against HB was greater than 50% and reached the highest, and on the 13thd, the bacteriostatic rate of strain 15 against HM reached the highest, but the difference was not much from that on the 12thd. Strain 19 reached the maximum bacteriostatic rate against both XF and FP on the 8thd, but had no obvious effect; on the 10thd, the bacteriostatic rate of strain 19 against HM reached a maximum (greater than 60%); on the 13thd, the bacteriostatic rate of strain 19 against HB reached the highest level (greater than 6%), and the bacteriostatic rate did not change much compared with that in 11-12 d; the bacteriostatic rate suddenly decreased in 10-11 d and stabilized at the end, which is possibly related to the mycelium growing close to the treatment inoculation strain point, which needs further experimental study. The bacteriostatic rate reaching the highest value on different days may be related to the growth rate of mycelium or the interaction between bacteria, indicating that both strain 15 and strain 19 have inhibitory effects on HB and HM and have research significance. Strain 19 has the strongest inhibitory effect on HM, while its optimization needs further study.
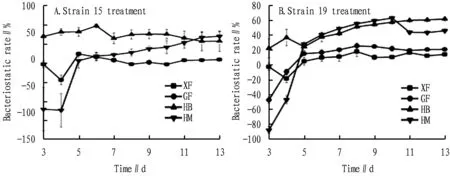
Fig.4 Variation trend of bacteriostatic rate of different strain treatments in 3-13 d
3.4 Bacteriostatic activity of strain 15 and strain 19The bacteriostatic activities of different strains against four diseases of ginseng are shown in Table 1.
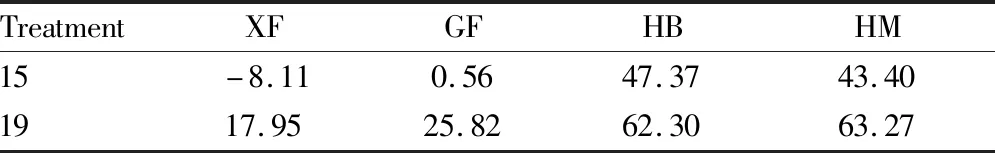
Table 1 bacteriostatic activities of different strains against four diseases of ginseng %
3.5 Determination of the growth cycle of strain 19From Section3.4, it can be seen that strain 19 has the best bacteriostatic effect on ginseng grey mould, and theOD600of the bacterial solution is positively correlated with the actual number of viable bacteria in the solution. Then, we determined theOD600value of strain 19 within 24 h and plotted the growth trend curve of the bacteria (Fig.6A). The seed solution entered the logarithmic growth phase about 8 h after inoculation, and the number of viable bacteria in the culture medium tended to be stable for 18-22 h. we selected bacterial solution with differentOD600values at 0, 6, 10, 14, 22 and 24 h for filter paper test.

Fig.5 Inhibition of strain 19 against HM
3.6 Bacteriostatic effect of differentOD600values of culture medium on ginsengB.cinereaThe inhibitory effect of differentOD600values of culture medium of strain 19 on ginsengB.cinereais shown in Fig.6B. The bacteriostatic activity of the extract within 24 h increased with the increase ofOD600value, and theOD600value in the range of 0.1-1.0 was positively correlated with the bacteriostatic rate. Its regression equation isy=19.876x+51.932,R2=0.926 2. WhenOD600>1, there is no linear relationship. Therefore, whenOD600<1, it is able to accurately read the corresponding bacteriostatic rate from the table. WhenOD600is in the range of 0.8-1.0, the bacteriostatic effect is the best, as high as 70%. The bacteriostatic rate shows a trend of increasing first and then decreasing, which may be related to the activity of bacteria. When the concentration of the culture solution is too large (the culture time is too long), a large number of dead bacteria in the culture solution will affect the accuracy of the measurement[15].
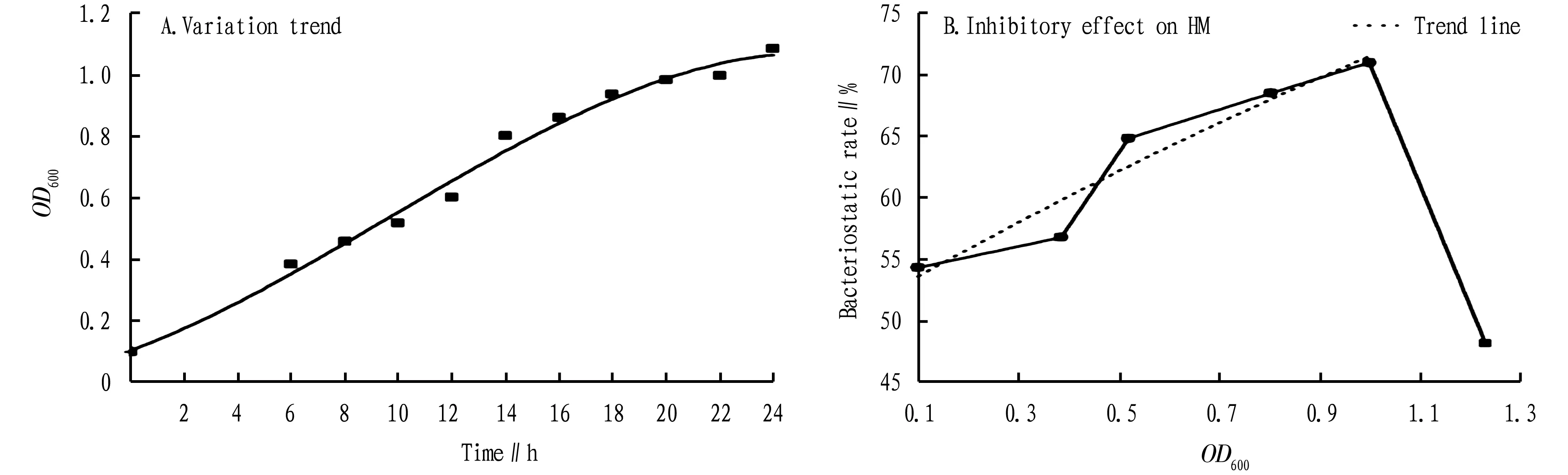
Fig.6 Variation trend of OD600 of culture medium with time and inhibitory effect on HM during liquid culture of strain 19
4 Discussion and conclusions
As a type of biocontrol bacteria and rhizosphere growth-promoting bacteria with application value,SphingomonasandPseudomonashave been studied more in the prevention and control of fruit and vegetable diseases[16-17]. However, there are few reports about their role in prevention of ginseng diseases. In this experiment, we conducted an indoor bacteriostatic test ofSphingomonasandPseudomonasagainst 4 species of pathogenic bacteria of ginseng diseases. The experimental results show that the growth of strain 15 and strain 19 tended to be stable in 8-10 d. The antagonistic ability of both bacteria against XF and GF was not strong, both below 30%, while the bacteriostatic rates against HB and HM were relatively high, ranging from 40% to 65%. Strain 19 had the strongest bacteriostatic rate of 63.27% against HM, which was higher than the result of Xu Jiao[18].
Because the strain 19 had the highest bacteriostatic rate against HM, we selected the strain 19 for growth cycle determination, and carried out the filter paper test with the seed solution with differentOD600values. In the research process of this bacterium, hemocytometer and plate colony counting method are commonly used in the measurement of bacterial suspension concentration, but both have their own shortcomings. The former has large errors, while the latter is complex and time-consuming. A large number of experiments have proved that the relationship between the concentration of different bacteria suspensions and theODvalue is specific[19-20]. In this experiment, we further studied the growth curve ofP.aeruginosaand the relationship between the bacteriostatic rate ofP.aeruginosaagainst HM and the turbidity (OD600) measured by spectrophotometer, and obtained the regression equation. Further experiment indicates that the bacteriostatic activity of the extract was enhanced with the increase ofOD600value, and theOD600value was in the range of 0-1. Its regression equation isy=19.876x+51.932,R2=0.926 2. From the regression equation, it can be seen that theOD600value is positively correlated with the bacteriostatic rate, and the correlation degree is high.OD600value in the range of 0.8-1.0 has the best bacteriostatic effect, up to 70%. WhenOD600>1, there is no linear correlation, which may be related to the activity of bacteria. When the concentration of the culture solution is too large (the culture time is too long), a large number of dead bacteria in the culture solution will influence the measurement accuracy[15]. Therefore, when using UV spectrophotometry to measure theOD600value of the bacterial solution, it should be carried out within a certain culture concentration and culture time.
This experiment provides convenience for more effective inoculation, establishes a fast, simple and accurate method for the determination of the best bacteriostatic rate ofP.aeruginosaculture solution to HM, and lays a foundation for large-scale culture ofP.aeruginosaculture solution. Besides, it is expected to provide a theoretical basis for the efficient control of ginsengB.cinereain field production, use it for the prevention and control of ginseng shoot diseases, and provide a reference for the efficient and diverse development of biocontrol agents for ginseng shoot diseases. In future, it is necessary to further study the control effect and mechanism ofP.aeruginosain ginseng fields, and whether it has any effect on the growth of plants.
 Asian Agricultural Research2022年5期
Asian Agricultural Research2022年5期
- Asian Agricultural Research的其它文章
- Status Investigation and Countermeasures of Green Development of Black Talc Industry Chain in Guangfeng District, Jiangxi Province
- Cloning and Bioinformatics Analysis of TpiA Gene of Vibrio alginolyticus HY9901
- Inhibitory Effect of Lactic Acid Bacteria on Salmonella
- Bioinformatics Analysis of Xylanase xynMF13-GH10 Gene from Mangrove Fungi
- Methodological Research in the Field of Civil Aviation Safety
- Safety Evaluation of Water Environment Carrying Capacity of Five Cities in Ningxia Based on Ecological Footprint of Water Resources
