PATHOGENICITY AND GENOMIC ANALYSES OF DENGUE VIRUS UNDER POSITIVE SELECTION IN HUMAN AND MOSQUITO CELLS*
ZHU Xiao-juan JIANG Yu-ting ZHANG Heng-duan GAO Jian WU Zhi-ming LI Chun-Xiao DONG Yan-de XING Dan GUO Xiao-xia ZHAO Tong-yan
(Beijing Key Laboratory of Vector Borne and Natural Infectious Disease, State Key Laboratory of Pathogens and Biosecurity, Institute of Microbiology and Epidemiology, AMMS, Beijing 100071, China)
Abstract Dengue virus (DENV) perpetuates in an alternating cycle replication in arthropod and vertebrate hosts. It is particularly important to gain comprehensive understanding of host-virus interactions and the relationship between genetic variation and virulence. To simulate the transmit of DENN between human hosts and mosquito vectors, DENV was repeatedly passaged between human hepatocellular carcinoma cell line (Huh-7) and mosquito cell line (C6/36). After 10 and 18 passages, the fitness of viral populations was measured with one-step growth curves and their virulence to mice and vector transmissibility were evaluated. Mutations in each population were determined by nucleotide sequencing. Viral fitness of DENV-2 decreased following alternating passages but increased after serial passages. Analysis of the amino acid sequences revealed that DENV that underwent continuous passage in C6/36 cells accumulated fewer mutations than virus passaged in Huh-7 cells or transferred between both cell types. Furthermore, the increased virulence that observed in serial passages might be due to the amino acid changes at positions 155 and 186 of protein E and the decreased virulence that observed in alternate passages might be due to the mutation at position 307. These results explored relationship between pathogenicity and genomic mutations of DENN and are valuable to guide the monitoring of future outbreaks.
Key words Dengue-2 virus;Alternating passage;Nucleotide mutations;Virulence
Dengue virus (DENV) (Flaviviridae: Flavivirus) is the most important human arboviral pathogens. It causes the mild febrile illness known as dengue fever (DF) but also the much more severe dengue hemorrhagic fever/dengue shock syndrome (DHF/DSS). DENV is transmitted to humans byAedesmosquitoes in many tropical and sub-tropical countries (Gubler, 1998). The Global Burden of Disease study in 2013 reported that there to be 390 million dengue infections per year, of which 96 million manifest apparently, which is more than three times WHO′s 2012 estimate (Bhattetal., 2013). For now, a tetravalent, recombinant live attenuated vaccine, CYD-TDV, which was developed by Sanofi-Pasteur, was licensed in 19 countries and some other DENV vaccines were in Phase 3 trials (Ahmadetal., 2019). However, there are multiple issues and controversies about CYD-TDV. Reports showed that the administration of CYD-TDV is risky to seronegative vaccine recipients (Sridharetal., 2018) and WHO recommended that the vaccine be administered only to individuals who had a prior dengue infection.
DENV is a single-stranded, positive sense RNA virus of approximately 10.7 kb nucleotides in size that encodes three structural (core (C), envelope (E), and membrane (M)) and seven non-structural (NS) proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) (Wilder-Smithetal., 2019). About one nucleotide mutation per genome per replication arises due to the high error rate of the viral polymerase of DENV (Coffeyetal., 2011). Quasispecies arise from rapid genomic evolution powered by the high mutation rate of RNA viral replication (Domingo, 2000; Vignuzzietal., 2006; Ciotaetal., 2010). The requirements of the internal population structure of a viral quasispecies to adapt to a changing environment depend on the nature of the selective constraint confronted by the viral population. Phylogenetic studies suggest that the rate of DENV evolution has accelerated substantially, as well as the disease severity (virulence), most likely driven by the surge in transmission (Rothmanetal., 1999; Aaskovetal., 2006; Vignuzzietal., 2006; 2008). However, the relationships between virulence and DENV genetic diversity are poorly understood.
Previous studies indicate that the emergence of pathogenic RNA virus is related to their genomic diversity and changes of their hosts (Drake, 1993; Domingo, 2000; Wangetal., 2000; Crottyetal., 2002; Coffeyetal., 2011). The increase in genetic diversity could have important phenotypic implications, such as the emergence of populations with altered antigenicity, or virulence (Twiddyetal., 2003; Vasilakisetal., 2009). Therefore, it is important that increasing genetic variation may facilitate even more efficient circulation and persistence of DENV in endemic areas, further increasing the risk of severe epidemics. To gain a better understanding of the relationship between genetic variation and pathogenicity of DENV in the presence of selective pressure, particularly given its reliance on both mosquitoes and mammals for transmission, we used Huh-7 and C6/36 cell lines as surrogate hosts for studying DENV host range evolution, and tried to determine a correlation of nucleotide changes with virulence or avirulence.
1 Materials and Methods
1.1 Mosquito
Aedesaegypti(Haikou, China), used in experimental infection, was previously collected from Haikou city of Hainan province, China, and reared under standard insectary conditions at (26 ± 1)℃ and (75 ± 5)% relative humidity, with a photoperiod of 14:10 (h) of light and dark cycles. Adult mosquitoes were provided with 8% sugar supplement until the infection experiment.
1.2 Mice
1-day-old suckling mice (Kunming mice) (Beijing Experimental Animal Center, Beijing, China) were used for all of the animal experiments. This study was carried out in strict accordance with the recommendations in the Guidance for the Care and Use of Laboratory Animals of the National Institutes of Health. All the experimental protocols were approved by the Laboratory Animal Center, State Key Laboratory of Pathogen and Biosecurity, Institute of Microbiology and Epidemiology IACUC’s (The permitted number is BIME 2011-09).
1.3 Virus and cells
DENV-2 (New Guinea C strain) was obtained from the Microbial Culture Collection Center of the Beijing Institute of Microbiology and Epidemiology (Beijing, China). The C6/36 cells were cultured in RPIM 1640 (Gibco, USA) culture medium, supplemented with 10% heat-inactivated FBS (Gibco, USA) and maintained at 28℃ without CO2. Huh-7 and Baby hamster kidney (BHK) cells were maintained in DMEM (Gibco, USA) with 10% heat-inactivated FBS, 1% penicillin and streptomycin (Gibco, USA), 0.5%L-Glutamine (Gibco, USA) and incubated at 37℃ with 5% CO2.
1.4 Virus passages
The parent DENV-2 strains were performed in different ways to gain three passage regimens: (i) serial, exclusive passage in Huh-7 cells; (ii) serial, exclusive passage in C6/36 cells; (iii) alternating passage between Huh-7 and C6/36 cells, with C6/36 cells being the ending cell line of each cycle. Each strain was passaged in each regimen in duplicate for a total of eighteen passages. Viruses were diluted before the first passage to yield at a multiplicity of infection (MOI) of 0.01, which resulted an 80% CPE. Infected cell culture supernatants were harvested on 4~5 dpi (day post infection) and pooled together before filtered through a 0.22 μm filter (Millipore, Germany) to achieve the maximum viral titer. The supernatants were then frozen at -70℃ until used to infect the next passage at 0.01 MOI.
1.5 Virus titration
The yield cell supernatants were quantified by a plaque assay on BHK cells as following: Ten-fold serial dilutions of supernatants containing DENV-2 viruses with DMEM, supplemented with 2% FBS, were added to confluent BHK cells monolayers grown on 12-well plates (Costar, USA). Virus adsorption was continued for 2 hours in a humidified incubator at 37℃ with 5% CO2supplement. The plates were rocked every 15 min during the 2-hour incubation. Every well was overlaid with 1.5 mL mixture of 2×DMEM (Gibco, USA) containing 4% FBS, and 2% liquefied ultrapure agarose (1∶1), and then incubated undisturbedly for 6 days. Cell monolayers were, subsequently, fixed with 4% formalin for 30 min, followed by being stained with 1% gentian violet dissolving in 20% alcohol for 30 min. Viral titers were calculated as plaque numbers times dilution rate.
1.6 Viral growth kinetics
The replication kinetics of DENV-2 mutants, which were originated from the parent virus (Parent), and the 10th, 18th passages (P10, P10-Alt, P18, P18-Alt) for each passage regimen, were generated and analyzed as follows: confluent monolayers of C6/36 or Huh-7 cells attached to 6-well plates were infected with different DENV-2 populations at a MOI of 0.01, in duplicate, and incubated in a chamber at 37℃ for 2 hours until the inocula were replaced with 2.5 mL maintenance medium. Virus samples from individual wells were harvested and purified by low speed centrifugation at 1, 2, 3, 4, 5 and 6 dpi, respectively, and quantified via qRT-PCR.
1.7 qRT-PCR
Viral RNA was extracted with Trizol (Invitrogen, USA) and quantified with TaqMan AgPath-IDTMOne-Step RT-PCR Kit (Applied Biosystems, USA) using ABI prism 7500 Real Time PCR System. Sequence-specific primers and nested TaqMan probe were derived from the conservation sequence of DENV-2 envelope (E) region and designed with Primer Premier 5.0 software (PREMIER Biosoft International, USA) as follows: a sense primer (nt1 166-1 189): 5’-AAGGAGAACCCAGCCTAAATGAA-3’, an antisense primer (nt1 270-1 293): 5’-GAACATAGCACAGGTCACAATGC-3’ and the probe (nt1 204-1 231): 5’-TTCGTCTGCAAACACTCCATGGTGGAC-3’. The TaqMan probe was labeled with a FAM at the 5’ end and a TAMRA at the quencher 3’ end. The RT-PCR cycling parameters were set as follows: the RT reaction at 45℃, 10 min; 95℃, 10 min; and followed by 40 cycles of PCR reactions at 95℃, 15 s, and 58℃, 45 s. The copy numbers of DENV-2 RNA were calculated by the standard curve method using serial dilutions of known amount of plasmid containing fragment of DENV-2 E gene.
1.8 Vector susceptibility
Vector susceptibility of above DENV-2 mutants was determined by oral infection ofAe.aegypti. The mouse blood meal, mixed with virus suspension and 8% sucrose solution (1∶1∶1), was warmed to 37℃ and fed the mosquitos using a Hemotek membrane feeding system housed in a feeding chamber. Fully engorged mosquitoes were maintained in an insectary at 29 ± 1℃ and (80 ± 5)% relative humidity under a photoperiod of 8:16 h (light∶dark) for 10 days, until mosquitoes were incapacitated for the following detection: heads and legs were collected in individual tubes to determine the viral dissemination rate while bodies were assayed to determine the mosquito infection rate via qRT-PCR.
1.9 Pathogenicity of different DENV-2 mutants
To assess the pathogenicity of different DENV-2 passage regimes, 20 μL volume of viruses were injected into the cerebrum of 1-day-old mice and symptoms such as convulsions and hunched posture were monitored. For median lethal dose (LD50) determination, eight groups of 32 1-day-old mice were intracerebrally infected with 20 μL of successive 10-fold diluted DENV-2. Survival was monitored three times a day for a total of 20 days. Calculations of LD50were done using the Reed-Muench formula.
1.10 Genomic sequencing of different DENV-2 populations
To investigate potential determinants of virulence, the genomes of the parent, 10th and 18th passage populations were analyzed. Viral RNA of passaged DENV-2 was extracted using Trizol (Invitrogen, USA) according to the manufacturer′s instructions. cDNA was synthesized using SMART MMLV Reverse Transcriptase (Takara, Cat. 639522) at 45℃ for 30 min, followed by 35 rounds of amplification with PrimeSTAR Max DNA Polymerase (Takara, Cat. R045) in a 50 μL reaction volume. The PCR products were purified and sequenced commercially by the Bejing Genomics Institute (BGI).
1.11 3 D Protein structure graphics of parent DENV-2 and mutants
The dengue virus structure was derived with the Swiss-Model structure prediction server via Expasy (www.expasy.ch) and drawn with Swiss-PDB Viewer. The three-dimensional (3 D) structure of a soluble ectodomain fragment of the dengue virus envelope protein was used as the basis for all depictions of E protein and models of mutants. The software program PyMOL v1.6 was used for protein structure graphics and the Homology module of this program was used for modeling loops and side chain rotamers.
1.12 Statistical analysis
Plaque diameter of different groups were compared using Brown-Forsythe and Welch ANOVA tests with Dunnett′s T3 multiple comparisons test. The replication dynamics of viral populations in Huh-7 or C6/36 cells were compared using a repeated-measure ANOVA on the triplicate titer values of each population at each time point, followed by a Tukey′s post-hoc test to detect differences among passage regimens. The chi-square test was used to compare the difference of infection rates or dissemination rates between different groups. Statistics were performed using Microsoft Office, the SPSS or Graph Pad Prism version 8.
2 Results
2.1 Plaque morphology of DENV-2 after serial or alternating passage
Viral populations, serial passage in Huh-7 or C6/36 cells, and alternating passage between both cell types, were obtained after ten or eighteen DENV-2 passages (P10, P18) from each passage regimens. The plaque assay on BHK cells showed that the 10th and 18th serial passages restricted to Huh-7 or C6/36 cells displayed a marked increase in plaque size while 10th and 18th alternating passages between Huh-7 and C6/36 cells declined relative to the parental strain (Fig.1), which indicated that DENV-2 gained a higher invasion rate after the serial passage than the alternating passage.
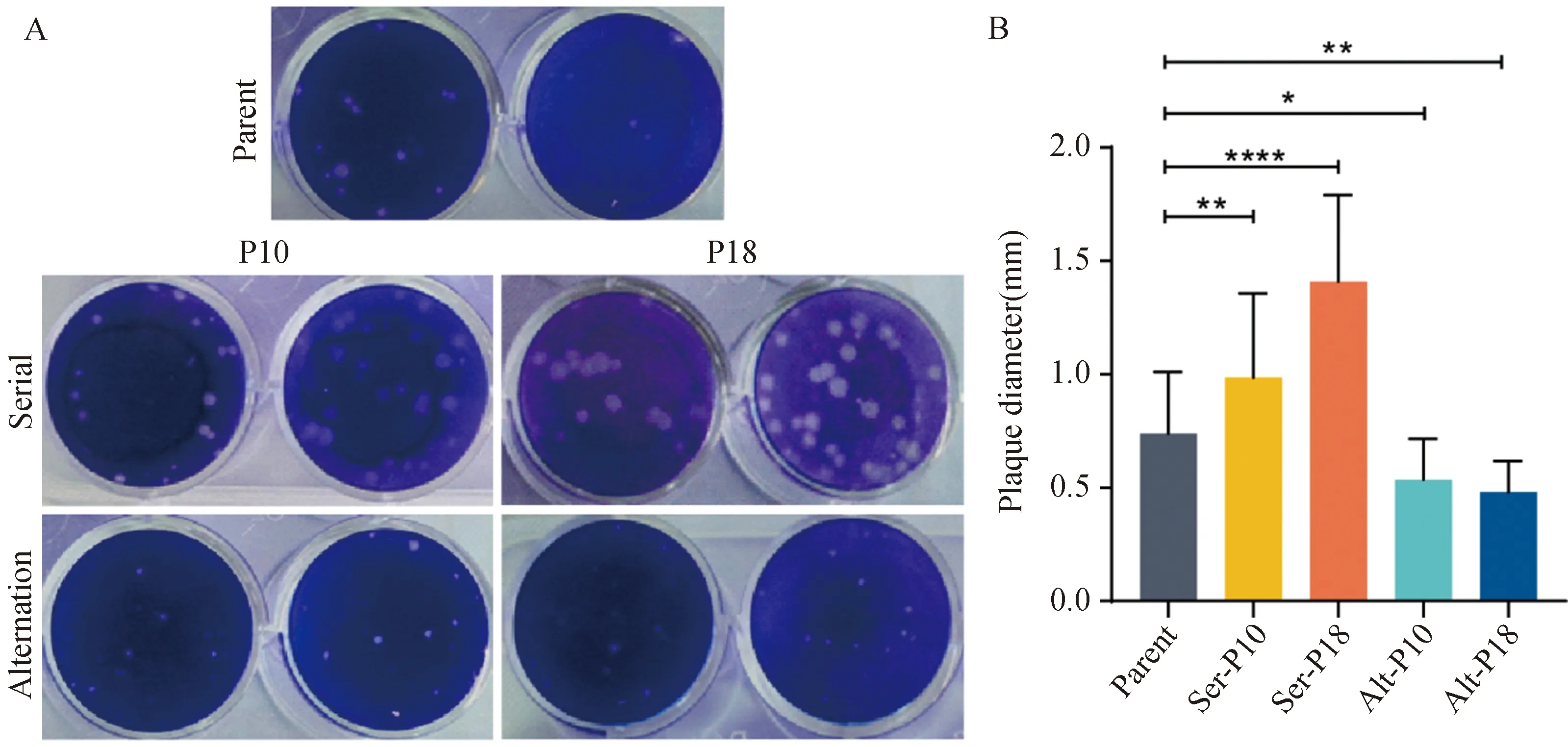
Fig.1 The plaque morphology of different DENV-2 populations
2.2 The virus replication dynamics of DENV-2 after serial or alternating passage
To determine how serial or alternating passage associates with DENV-2tness, analysis of virus replication comparing passage 10, passage 18 (P10, P18) and parent populations was conducted in C6/36 and Huh-7 cells, respectively. When replication was assessed based on viral RNA yields, strikingly different results were observed between mosquito cell and primate cell (Fig.2). In C6/36 mosquito cells, the serial passage populations (P10/18-C6/36, P10/18-Huh-7) resulted in an increasing replication kinetics curve in the later phase of the infection (3-6 dpi) relative to the parent population (Fig.2-A). However, the replication kinetics curve of the alternating passage populations (P10/18-Alt) was lower than that of the parent (Fig.2-B).
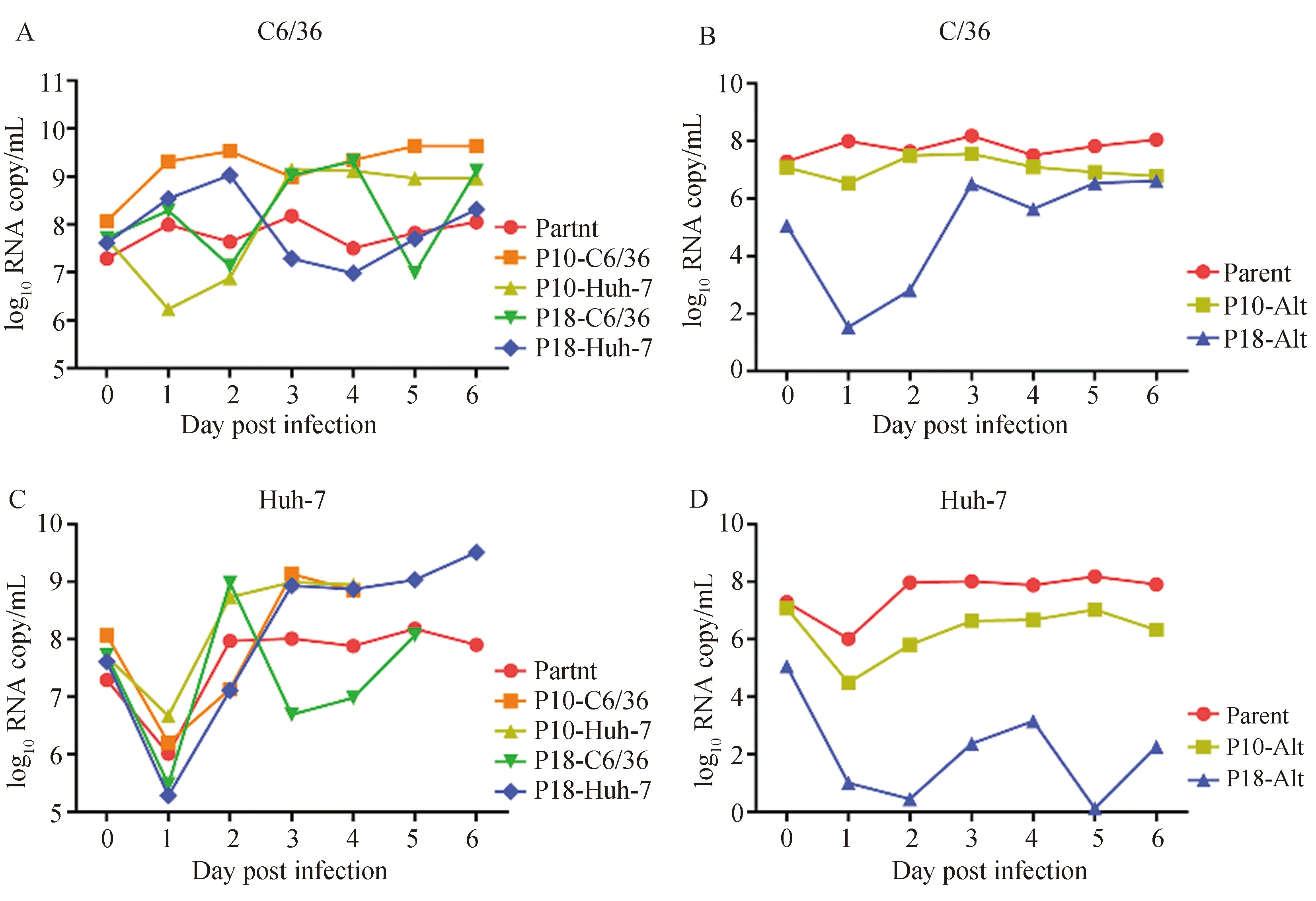
Fig.2 Determination of virus replication dynamics
When their replication was evaluated in primate Huh-7 cells, all viral RNA of the serial passage and parental populations were reduced at 1 dpi compared to 0 dpi, but they were increased at 2 dpi until 6 dpi (Fig.2-C). The alternating passage populations showed much lower RNA copies than the parental and exhibited low replication from 1 to 6 dpi after 18 alternating passages (Fig.2-D). Intriguingly, we noticed that DENV-2 populations after 18 serial passages (P18-C6/36 and P18-Huh-7) showed a higher replication level since 3 dpi in the cell line in which the virus was passaged but a lower replication level in the other (Fig.2-A, C).
2.3 Assessment of pathogenicity and determin-ation of median lethal dose for mice
1-day-old suckling mice were used to evaluate the virulence of DENV-2 undergone exclusive or alternating passaging. Results showed that parental virus-infected mice exhibited symptoms on day 7 post infection, while mice that were inoculated with exclusive passaged virus developed symptoms on 6-7 days and no symptoms were observed until 9-12 days on the mice inoculated with alternating passaged virus. Those observations were coincident with previous results that the pathogenicity of DENV-2 increased in serial passage and decreased in alternating passage. To further assess the pathogenicity of different virus mutants, the LD50of parental, P10-Alt and P18-Alt populations to suckling mice were measured. As shown in Tab.1, parental virus exhibited 104.5LD50/20 μL to suckling mice, which is around 148 (102.17) times of P10-Alt population and 676 (102.83) times of P18-Alt population.
Tab.1 LD50 measurement of parental, 10th and 18th alternating passages of DENV-2 populations
2.4 Susceptibility in Ae. aegypti
To further assess the susceptibility of different virus mutants to the vector, infection and dissemination rates of those mutants toAe.aegyptiwere determined. Results showed that there are no significant differences in infection rates among these mutants and parent virus. However, P18-Huh-7 exhibited significantly lower dissemination rate compared to the parent virus (P<0.01) or P18-C6/36 (P<0.05) (Tab. 2). This was reminiscent of the low replication level of P18-Huh-7 compared to the P18-C6/36 at 3, 4 or 6 dpi in mosquito cell line (Fig.2-A).
Tab.2 DENV-2 infection and dissemination rates in Ae. aegypti
2.5 Variations of DENV-2 after serial or alternating passage
Based on the above analyses, the 10th and 18th passage populations were chosen to assess the intra-population genetic diversity in the genome. After 10 alternating passages, DENV-2 acquired 6 consensus mutations throughout the genome, 5 of which, E-6 (953), E-307 (1856), NS2A-33 (3576),NS4B-238 (7538) and NS5-831 (10062), were non-synonymous. When the same strain specialized in Huh-7 cells, it accrued 7 consensus mutations, 5 non-synonymous mutations of which distributed in M-119 (794), E-6 (953), NS2A-33 (3576), NS4B-238 (7538) and NS5-831 (10062). Similarly, C6/36-passage 10 virus accrued 5 consensus mutations, 3 of which were non-synonymous and distributed in E-6 (953), E-155 (1400) and NS2A-38 (3589) (Tab.3).
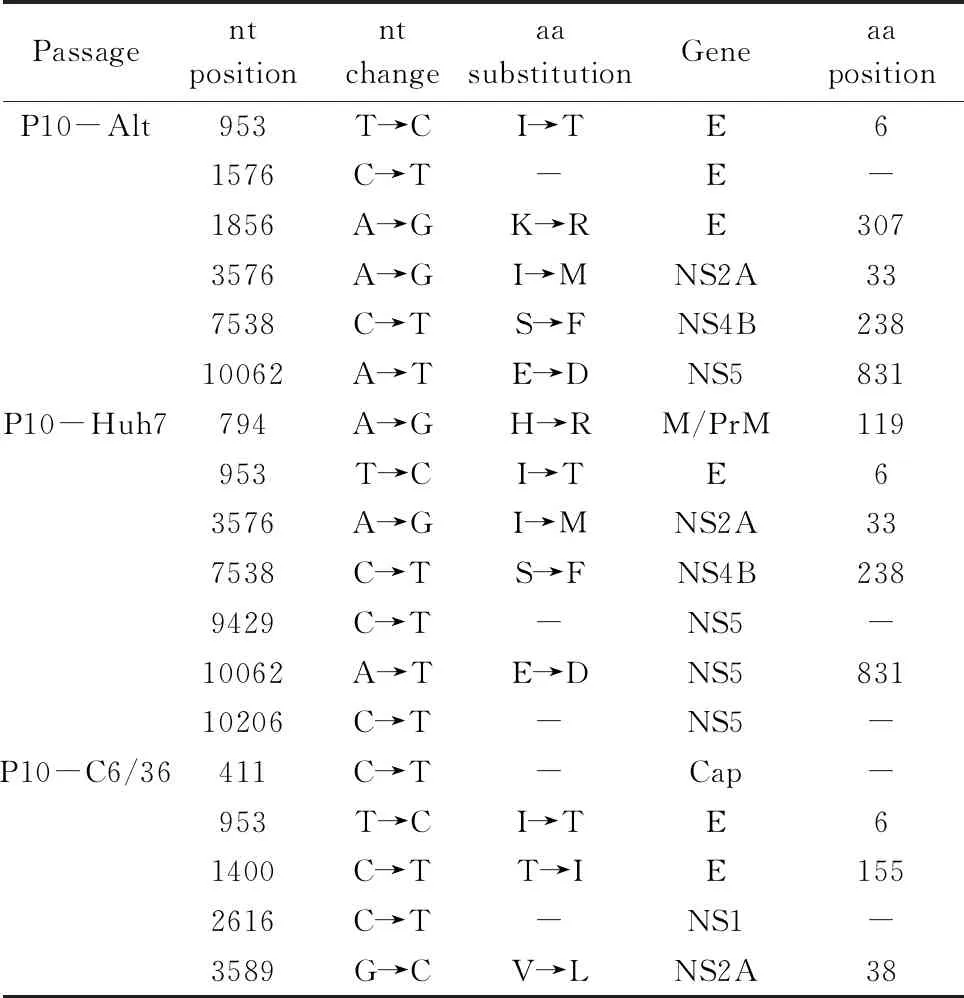
Tab.3 Nucleotide (nt) and amino acid (aa) sequence changes of passage 10 DENV-2 relative to the parental population
Successive 10-fold diluted DENV-2 populations were intracerebrally injected to the 1-day-old mice and survival was monitored to determine the median lethal dose using the Reed-Muench formula.
Furthermore, after 18 alternating passages, a total of 8 convergent mutations were observed, 5 of which were non-synonymous and distributed in E-6 (953), E-47 (1076), NS2A-33 (3576), NS4B-156 (7291) and NS4B-238(7538). A total of 13 consensus mutations accrued after 18 passages in Huh-7 cells, 7 of which were non-synonymous and distributed in M-119 (794), E-6 (953), E-124 (1306), E-186 (1493), NS2A-33 (3576), NS4B-238 (7538) and NS5-831 (10062). In addition, 18 serial passages in C6/36 cells accrued 6 consensus mutations, 4 of which were non-synonymous and distributed in E-6 (953), E-124 (1306), E-293 (1814) and NS2A-33 (3576) (Tab.4). Over all, we noted that there were most observed ORF mutations of consensus sequences in Huh-7-passage 18 viruses and least in successive C6/36-passag viruses.
Tab.4 Nucleotide (nt) and amino acid (aa) sequence changes of passage 18 DENV-2 relative to the parental population
To estimate the possibility that mutations in alternatively passaged viruses were the result of bottleneck events occurring soon after the first transfer of virus between Huh-7 and C6/36 cells, virus derived from the 2nd passage was sequenced. The fact that only 1 consensus mutation was found at this early stage but 6 were found in virus from the 6th alternate passage (Tab.5) suggests that these mutations were not due to bottlenecks but developed over time, either stochastically, or due to positive selection.
Tab.5 Mutations of alternating passage 2 and 6 DENV-2 relative to the parental viruses
2.6 3 D structure of the parental and mutant DENV-2 E protein.
According to the above investigations of DENV-2 mutations after different passage procedure, all 7 non-synonymous mutations in the E gene, which resulted in I6T, K47R, N124Y, T155I, S186F,Q293R and K307R acid amino substitution in the E protein, were selected for our study. Based on the crystal structure of tick-borne encephalitis virus E protein, available crystallographic models of the E ectodomain and atomic structure of DENV E protein (PDB ID: 3J27 A), a predicted structure of the parent DENV-2 E protein was mapped (Fig.3-A). Seven amino acid substitutions were introduced to the model and constituted a new 3D model as shown in Fig.3-B. The topography of those substitution sites on the E protein dimer and a detailed view of our interesting region are shown. Those changes of atoms could have influences on the affinity between viruses and host cells.
Fig.3 Views of a predicted structure of the dengue virus E proteinThe predicted 3D structure of parental (A) or mutant (B) DENV-2 E protein. Amino acid residues of 6, 47, 124, 155, 186, 293 and 307 were indicated in the view. Atom colors: green, carbon; blue, nitrogen; red, oxygen.
3 Discussion
In the present study, the alternating invertebrate-to-vertebrate host cell passage model was served as aninvitrosystem to predict the relationship between genetic changes and virulence. DENV-2 strain was passaged 10 or 18 times, either continuously or alternately in Huh-7 and C6/36 cells. Three assays were used to evaluate virulence of those virus populations. Firstly, the fitness change of mutants was evaluated by measuring virus replications in C6/36 and Huh-7 cells respectively. Secondly, the virulence of serial and alternate passage of DENV-2 to mice was compared, and finally, viral transmissibility in vector mosquitoes was determined. Taken together, the results showed that low-passage strains of exclusively passed virus (P10-Huh-7 and P10-C6/36) exhibited higher replication levels compared to the parent in the mammalian or mosquito cells infection but high-passage strains (P18-Huh-7 and P18-C6/36) reached a replication peak only in the cells in which they were passed. This is coincident to the conclusion made by Vasilakis (Vasilakisetal., 2009) that viruses allowed to specialize in single cell types exhibited fitness gains in the host cell but fitness losses in the bypassed cell type. It also explains why P18-Huh-7 showed low dissemination rate inAe.aegypti. However, the alternately passed virus (P10/18-Alt) demonstrated small plaque size and low replication curve in cells infection, weak virulence to the sucking mice and relative low infection rates to the mosquitoes, which means DENV-2 showed fitness losses during alternating passage. This is opposite to the Vasilakis′study in which most alternating passages exhibited fitness gains in both cell types (Vasilakisetal., 2009). But in their study, the virus only passed 10 cycles alternately between C6/36 and Huh-7. More cycles of alternate passage may contribute to virulence attenuation.
Next, viruses were assayed genetically by sequencing of the genome to explore the relationship between genetic change and virulence. Our study showed that DENV that underwent continuous passage in C6/36 cells accumulated fewer mutations than virus passaged in Huh-7 cells and virus alternately passaged in both cell types. It can be explained that the C6/36 cell line has a dysfunctional antiviral RNA interference system (Brackneyetal., 2010) and holds a weaker positive selection environment compared to Huh-7 cell line. In this study, the vast majority of nucleotide substitutions were non-synonymous, which contrasted sharply with arbovirus mutations that occur in nature, where synonymous mutations vastly outnumber non-synonymous mutations, suggesting purifying selection (Jerzaketal., 2005; Lequimeetal., 2016). The ratio of non-synonymous (dN) to synonymous (dS) substitutions in this study is 2, suggesting convergent evolution via positive selection.
The E protein is essential to DENV virulence, and is responsible for host cell attachment, entry, viral-endosome membrane fusion, and virus assembly (Butrapetetal., 2011). According to the above analyses, combined with the model representing mutant E, we found that the substitutions of T155I, S186F and K307R may affect virulence. Kawanoetal. (1993) compared acid amino sequence between nonneurovirulent DENV-4 strain 814669 and mouse-neurovirulent DENV-4 strain H241. They found that three amino acid differences located in the E protein, and the single substitution T155I contributed most to the neurovirulence of strain H241, which ablated one of the two conserved glycosylation sites. Butrapetetal.(2011) reported that four DENV-2 E protein molecular hinges are highly conserved in amino acid residues, and E-186 is located in the H3 region (186-196). When a mutant L135W DENV-2 grew in K562 cells, it evolved additional mutations S186F and N276N. These two mutations were regarded as compensatory mutations, because residues E-135, 186, 276 are close in space and essential to maintain the spatial structure of E protein. In our study, substitution S186F was observed in Huh-7-passage 18 (P18-Huh-7) DENV-2, which exhibited virulence increase in infection of Huh-7 cell type andAedesaegyptimosquitos. Similar results were seen in Chen′s work (Chenetal., 2003), in which, an S186F mutation (Although the author stated it′s an S186P mutation in their paper, we believed it’s a typo because it is the same nucleotide mutation in the same position as seen in our study.) of E protein occurred in DENV-2 that serially transferred in Vero cells after 20 or 30 passages and titers of those variants elevated 1-2 orders of magnitude in comparison with the parent virus on day 5 after reinoculated into Vero cells. E-307 is located on the upper surface of Domain III (Beasleyetal., 2002). It has been suggested that Domain III contains residues responsible for the determination of host range, tropism and virulence amongaviviruses (Ciotaetal., 2007). In particular, the structural Domain III of E protein has been proposed as a putative receptor-binding domain (Beasleyetal., 2002). Although neutralizing epitopes have been identified in other domains of the E protein, mutation K307R may be the most efficient at blocking the ability of viruses to attach to host cells, which can obviously reduce their virulence. Ourndings supported the idea that certain specific mutations of residues within Domain III can affect the virulence of flaviviruses (Barrettetal., 1990; Butrapetetal., 2000), and Beasleyetal.(2002) reported a Lys amino acid mutation at E-307 of WNV in MAbR variants associated with a slight attenuation of viral virulence.
Besides the sites of E-155, 186 and 307, other substitution sites through the genome may be also critical to pathogenesis and virulence of DENV-2. However, how these substitutions in DENV-2 affect the virulence and ability of binding to host cells needs further study. In summary, understanding the selective pressures that drive genetic and phenotypic changes in viruses is crucial to predict their ability to mutate and re-emerge, which in turn contributes to the development of effective ways of preventing or mitigating DENV infection.

