Molecular Cloning and Bioinformatics Analysis of crp Gene in Vibrio alginolyticus
Zhiqing WEI Weijie ZHANG Linlin YIN Haiyun FENG Junlin WANG Fuyuan ZENG Xing XIAO Huanying PANG
In the vast majority of microorganisms, the regulation of transcription initiation requires the participation of regulatory genes. Prokaryotes are more sensitive to changes in their external environment. In order to adapt to the complex and changeable external environment, prokaryotes will actively turn on or off the expression of certain genes [8]. Compared with eukaryotes, the transcriptional regulation mechanism of prokaryotes is relatively simple, and generally, regulatory proteins interact with RNA polymerase to activate the transcription process [9]. The crp gene encodes the cAMP receptor protein (CRP), which was first discovered in Escherichia coli catabolite repression, and was subsequently confirmed to be a bacterial transcriptional activator [9]. When the environment changes drastically, cAMP will activate CRP to form a complex, bind to a specific DNA site in the promoter region, and interact with RNA polymerase to promote transcription initiation [10]. Transcriptomic studies on E. coli found that CRP, as one of seven total regulatory proteins, is responsible for regulating about 50% of transcription initiation [11]. At present, the research on E. coli CRP is relatively in-depth, mainly focusing on carbon source metabolism. Studies have shown that CRP can positively regulate genes related to the tricarboxylic acid cycle (TCA) such as sdhA -D, sucA -D and fumA [12]; and it can directly regulate the transcriptional process of the succinate dehydrogenase complex sdhCDAB [13]. In addition to regulating carbon source metabolism-related genes, CRP can also directly or indirectly regulate the expression of other genes. CRP is a key regulator of Vibrio cholerae and can directly inhibit the expression of tcpP and the transcription of the outer membrane protein gene ompT [14]. Manneh et al. [15] found that CRP was related to the colonization ability of V. cholerae , and the infection experiment proved that the colonization ability of Δ crp was weakened compared with wild strains, and it could only colonize the upper intestine of zebrafish. In addition, the regulatory mechanism of CRP is also related to the virulence of bacteria. The virulence of Yersinia enterocolitica was weakened after the deletion of the crp gene, and the immunized mice had a certain immune protection against wild strains [16]. The fish immunized with Edwardsiella sp. J100Δcrp had high immune protection against wild virus challenge. The study of Li et al. [17] showed that compared with the parental strain, the virulence of Salmonella typhimurium deletion strain SL1344Δcrp to chicks decreased by about 99.99%, indicating that the deletion of the crp gene can significantly reduce the virulence.
The type III secretion system (T3SS) is a set of highly conserved pinhole-shaped virulence apparatus that penetrates the inner and outer membranes of bacteria, intercellular spaces and host cell membranes, and can inject virulence factors into host cells to interfere with normal physiological metabolism and lead to host death [18]. At present, it can be confirmed that T3SS is composed of effector proteins, apparatus proteins, transposon proteins, regulatory proteins and molecular chaperones. Regarding the relationship between CRP and T3SS, recent studies have shown that CRP can regulate the secretion of T3SS. YpkA and YopJ are effector proteins of T3SS and both are required for the virulence of Yersinia pestis [19-20]. CRP can specifically bind to the promoter region of the chaperone protein sycO and inhibit the expression of the sycO-YpkA-YopJ operon of Y. pestis of the Microtus fuscus type and affect its virulence. In addition, Zhan et al. [21] found that CRP might directly negatively regulate the transposon protein yopD through chip expression profiling and real-time quantitative RT-PCR. Tan [22] conducted bioinformatics analysis and predicted that CRP might affect the expression of Klebsiella pneumoniae type III secretion system through KP1-4563 gene transcription, thereby regulating bacterial virulence.
In recent years, there have been many studies on the crp genes of E. coli and Salmonella at home and abroad, but little has been done on the crp gene in V. alginolyticus . Therefore, in this study, the crp gene of V. alginolyticus was cloned, and its bioinformatics analysis was carried out, aiming to provide a reference for finding efficient protective antigens against vibriosis.
Materials and Methods
Materials
Strain and vector
The virulent strain of V. alginolyticus HY9901 was kept in our laboratory [23], and the cloning vector pMD18-T was purchased from Takara Company.
Main reagents
DNA polymerase was purchased from Takara company. Bacterial genome DNA extraction kit and DNA gel recovery kit were purchased from Tiangen company. Other reagents were imported or of domestic analytical grade. PCR primer synthesis and sequence determination were completed by Sangon Biotech (Shanghai) Co., Ltd. The concentration of ampicillin used was 100 μg/ml.
Experimental methods
Extraction of total DNA from V. alginolyticus
V. alginolyticus HY9901 was inoculated in TSB (2% NaCl) medium and cultured with shaking at 28 ℃ for 18 h. An appropriate amount of bacterial liquid was added into a centrifuge tube and centrifuged at 10 000 r/min for 1 min to collect bacterial cells, which were extracted for genomic DNA with a kit, and stored at -20 ℃.
Cloning of crp
A pair of primers were designed according to the complete gene sequence of V. alginolyticus registered on Genbank (ACCESSION: GU074526): the upstream primer F: ATGGTTCTAGGTAAAACCTCAA, and the downstream primer R: CCACGATGGTTTTTACCGT. The total DNA extracted from V. alginolyticus HY9901 was used as the template, and the PCR reaction conditions were as follows: pre-denaturation at 95 ℃ for 3 min, 32 cycles of denaturation at 95 ℃ for 30 s, annealing at 52 ℃ for 30 s and 72 ℃ for 60 s, and extension at 72 ℃ for 10 min. The PCR product was examined by 1.0% agarose gel electrophoresis and recovered by cutting the gel.
Sequencing of PCR product
According to the instructions, the PCR product was ligated to the pMD18-T vector, and then transformed into E. coli DH5α competent cells. Screening was performed on LB plates containing Amp+resistance, and positive clones were picked and sent to Guangzhou Shenggong Biotechnology Co., Ltd. for sequencing.
Bioinformatics analysis of the crp gene of V. alginolyticus
Referring to the method of Pang et al. [24], NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi) was used for sequence homology alignment and similarity analysis. ExPASy Proteomics Server (http://web.expasy.org/protparam/) was used to deduce the amino acid sequence, determine open reading frames (ORFs), and perform calculate molecular weight (Mw) and theoretical isoelectric point (pI) prediction. Signal peptide sequences were predicted by SignalP 4.0 Server (http://www.cbs.dtu.dk/services/SignalP). Transmembrane domains were predicted by TMHMM Server 2.0 (http://www.cbs.dtu.dk/services/TMHMM). SoftBerry-Psite (http://linux1.softberry.com/berry.phtml?Topic=psite&group=rograms&subgroup=proloc) was used to predict the distribution of functional sites in the amino acid sequence. Promoters were analyzed using Promoter 2.0 software. PSORT II Prediction (http://psort.hgc.jp/form2.html) was used to predict subcellular localization. A phylogenetic tree was constructed by the neighbor-joining method using Clastal 2.0 and MEGA 5.0 software. A three-dimensional structure was constructed using the SWISS-MODEL (http://www.swissmodel.expasy.org/) program. The pathways involved were analyzed by Kegg software (http://www.genome.jp/kegg/). The STRING database was searched for protein network interactions (http://string.embl.de/).
Results and Analysis
Cloning of crp
A specific band of about 633 bp was successfully amplified by PCR. The sequencing results showed that the crp gene contains an open reading frame (ORF) of 633 bp, encoding 210 amino acids. The accession number in GenBank is OM095380.
Physicochemical properties of CRP
The Vibrio CRP protein was analyzed by ExPASy software, and the results showed that the total number of atoms is 3 366, and the molecular structure is C1 052H1 706N290O308S10. The theoretical molecular weight is 23.665 4 kDa, and the theoretical pI value is 7.74. The instability coefficient is 29.65 (stable); the liposoluble coefficient is 97.95; the overall average hydrophilicity is -0.196; and the protein is overall hydrophilic. The protein does not contain selenocysteine (Sec) and pyrrolysine (Pyl), and the molar extinction coefficient at 280 nm is 18 575 (mol/cm). The total number of acidic amino acids (Asp+Glu) is 25; the total number of basic amino acids (Arg+Lys) is 26; and the N-terminal is methionine (Met). The half life of expression in yeast and E. coli is greater than 20 and 10 h, respectively, and the half life of expression in mammalian reticulocytes in vitro is 30 h.
Sequence analysis of CRP
The SignalP 5.0 Server program was used to predict the structure of the N-terminal signal peptide of the amino acid sequence of CRP, and it was found that there was no obvious signal peptide cleavage site and no signal peptide. The protein was predicted to have no transmembrane domain by TMHMM Server 2.0 program. The SoftBerry-Psite program predicted the amino acid sequence to have 3 protein kinase C phosphorylation sites (99-101aa, 129-131aa, 141-143aa), 3 casein kinase II phosphorylation sites (91-94aa, 159-162aa, 169-172aa), 1 N-terminal myristoylation site (174-179aa), 1 isoprenyl binding site (CAAXbox) (19-22aa), 3 microbody C-terminal target signal sites (19-21aa, 142-144aa, 185-187aa), 1 cyclic nucleotide binding domain 1 site (30-46aa); 1 cyclic nucleotide binding domain 2 sites (71-89aa), and 1 bacterial regulatory protein ( crp signature family) site (168-191aa). The prediction results of protein subcellular localization showed that the probability of CRP localization in cytoplasm, nucleus, vesicles, vacuoles and mitochondria of the secretion system was the same (33.33%).
The green parts represent the protein kinase C phosphorylation sites; the blue parts represents the casein kinase II phosphorylation sites; the purple part represents N-terminal myristoylation site; the red part represents the isoprenyl binding site; the black underlined parts represents the microbody C-terminal target signal sites; the red underlined part represents the cyclic nucleotide binding domain 1 site; and the blue underlined part represents the cyclic nucleotide binding domain 2 site.
Zhiqing WEI et al. Molecular Cloning and Bioinformatics Analysis of Gene crp in Vibrio alginolyticus
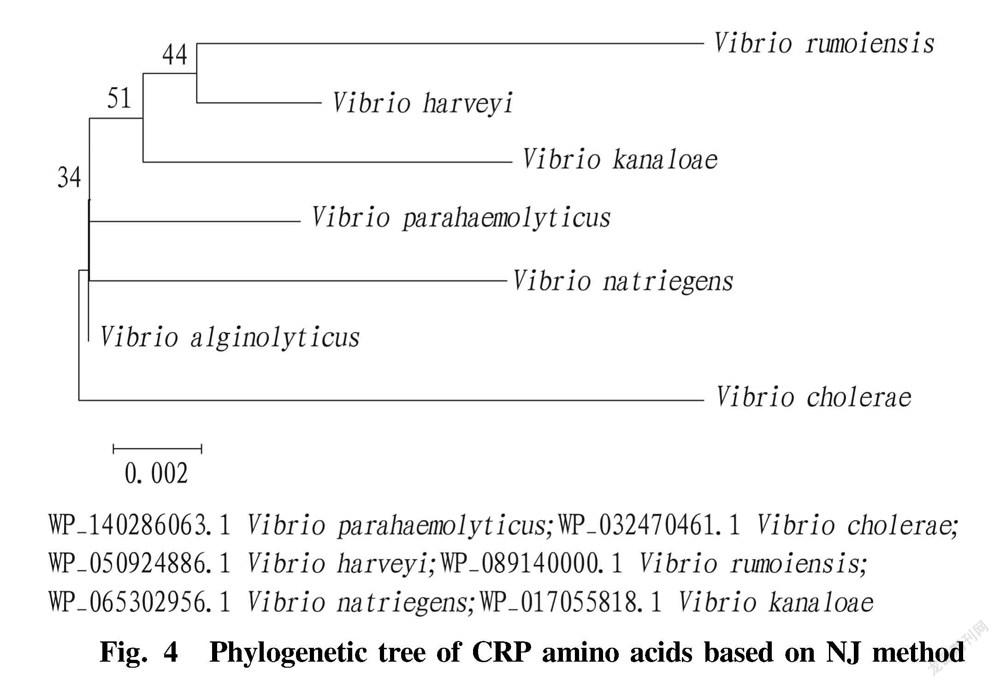
Homology and evolution analysis of CRP
The amino acid sequence homology alignment using NCBI showed that the protein encoded by this sequence is a general transcriptional regulator of a certain Vibrio that is activated by cAMP. BLAST analysis showed that the sequence has high homology with CRPs of other Vibrio species, and the homology with the CRP amino acid sequence of V. parahemolyticus is as high as 99.52%. The similarity comparisons of multiple sequences indicated that CRP is highly conserved in Vibrio .
A phylogenetic tree was constructed using the deduced CRP amino acid sequence of V. alginolyticus with Vibrio in the MEGA 5.0 software by the Neighbor-joining method. The results showed that the homology of CRP between V. alginolyticus and V. natriegens is high.
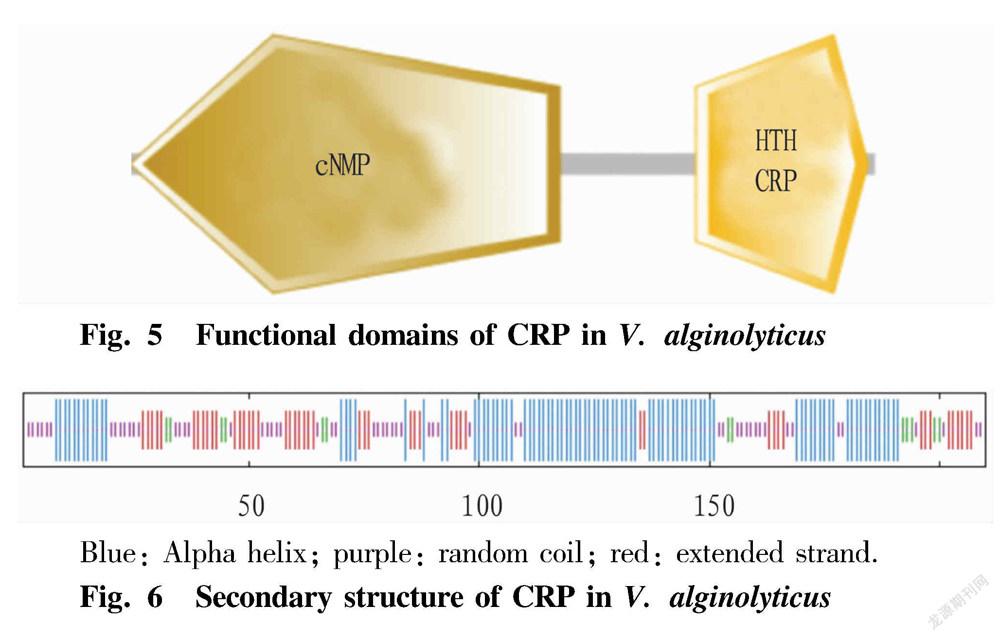
Functional domain and secondary structure prediction of CRP
The SMART program prediction showed that the protein has one cNMP domain (2-122aa) and one HTH CRP domain (160-208) (Fig. 5). In the secondary structure prediction, alpha helix accounts for 42.86%; random coil accounts for 27.14%; extended strand accounts for 23.33%; and β-sheet accounts for 6.67% (Fig. 6).
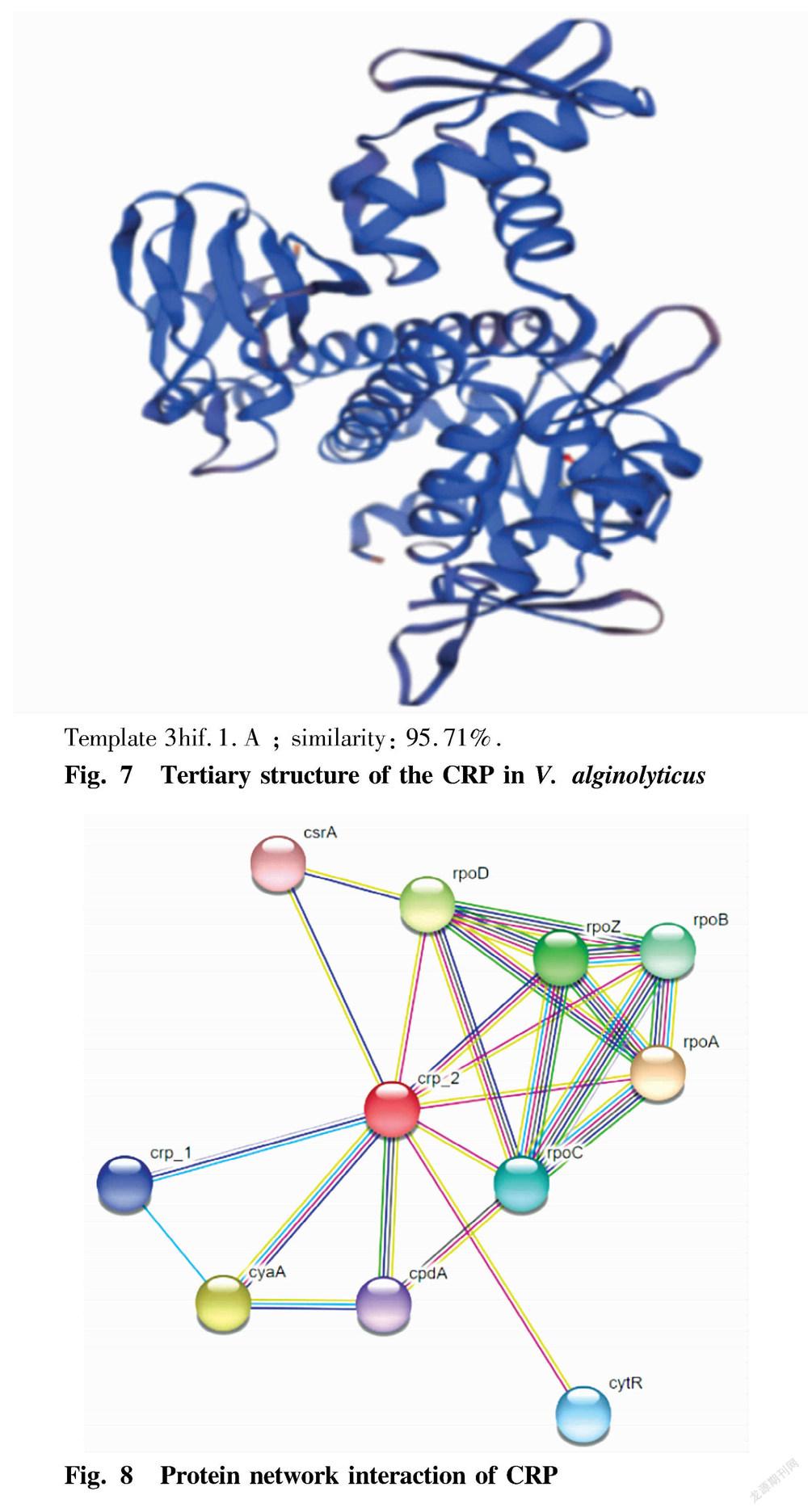
Tertiary structure prediction of CRP
The amino acid sequence of crp in V. alginolyticus was submitted to the SWISS-MODEL program, and the homologous protein was automatically searched as a template to obtain a single subunit tertiary structure model of CRP (Fig. 7).
Protein network interaction of CRP
In the protein network interaction, it could be found that the proteins interacting with CRPs include RpoD, RpoC, RpoZ, RpoB, RopA, GltB, CytR, VMC_01620, CyaA, and CpdA (Fig. 8).
Conclusions and Discussion
V. alginolyticus is one of the common pathogenic bacteria in the ocean, which has caused great economic losses to the aquaculture industry. At present, chemical drugs and antibiotics are mainly used in the prevention and treatment of the disease, and the resulting problems such as drug residues and increased drug tolerance in bacteria have become more and more serious [4]. Therefore, finding excellent protective antigens and developing new vaccines has gradually become one of the hot spots in aquatic disease research.
Bioinformatics analysis can predict the evolutionary relationship, physicochemical properties, secondary structure and tertiary structure of target proteins, and is thus an important technology for analyzing protein structure and function [25]. Lou et al. [26] used the DNAStar Protean software to comprehensively analyze the parameters including secondary structure, hydrophilicity and antigenic index of P1 protein, and successfully predicted the distribution of antigenic epitopes of its B cells. Petzke et al. [27] synthesized the B cell epitope of Sm25 molecule in Schistosoma mansoni combining biological software prediction and artificial methods, and it was found from ELISA test that the sensitivity and specificity of the epitope peptide were better than those of the control group. In this study, the crp gene of V. alginolyticus was successfully cloned and bioinformatic analysis was carried out. The results of sequence analysis showed that there was no obvious signal peptide cleavage site at the N-terminus of the CRP, and there was no signal peptide. It was inferred that the CRP is not a secreted protein. The analysis results of TMHMM Server 2.0 program prediction showed that the protein has no transmembrane domain. The SoftBerry-Psite program was used to analyze the amino acid sequence of the CRP protein, and it was deduced that it contains 3 protein kinase C phosphorylation sites, 3 casein kinase II phosphorylation sites, 1 N-terminal myristoylation site, 1 isoprenyl binding site, 3 microbody C-terminal target signaling sites, 1 cyclic nucleotide binding domain 1 site, and 1 cyclic nucleotide binding domain 2 sites. Protein phosphorylation can regulate proteins in real time, and is one of the general regulatory mechanisms for cells to respond to various stimulus signals [28]. Although V. alginolyticus lacks endoplasmic reticula and Golgi apparatuses, and cannot perform functions such as glycosylation and acylation of CRP proteins, it does not affect phosphorylation modification [23].
Through BLAST comparison on the protein sequences of various Vibrio , it was found that the CRP in Vibrio was highly similar, indicating that the protein is relatively stable during the evolution of Vibrio . The results of multiple-sequence analysis and alignment suggested that the function of CRP is relatively conserved, so it could be speculated that this protein may have important functions. The phylogenetic tree showed that the Vibrio CRP is closely related to V. natriegens .
In this study, the complete gene sequence of crp was successfully amplified from V. alginolyticus HY9901, and its bioinformatics analysis was carried out. The results of sequence analysis showed that the gene is 633 bp in full length, encoding 210 amino acids; the protein has the molecular structural formula C1051H1704N290O308S10, with a theoretical molecular weight of 23.6514 kDa, and has a theoretical pI value is 7.74, and it is stable and hydrophilic; and the CRP has no transmembrane domain, and has various sites. The prediction of protein subcellular localization showed that the CRP is most likely to be located in the cytoplasm, followed by the nucleus. The BLAST analysis found that the sequence has high homology with CRPs of other Vibrio species. The SMART program predicted the CRP to have one cNMP domain and one HTH CRP domain. The results of CRP secondary structure prediction showed that alpha helix accounts for 42.86%, random coil accounts for 27.14%; extended strand accounts for 23.33%; and β-sheet accounts for 6.67%. The similarity between its tertiary structure model and template 3hif.1.A is 95.71%.
References
[1] CHEN Q, YAN QP, MA S. Progress on pathogenicity research of Vibrio alginolyticus [J]. Marine Sciences, 2006(8): 83-89. (in Chinese).
[2] CHANG YS. Cloning and expression of vscD gene and study on the function of vscB gene in type III secretion system of Vibrio alginolyticus [D]. Zhanjiang: Guangdong Ocean University, 2018. (in Chinese).
[3] DING SY, HOU LY, YU CY, et al. Research progress on pathogenic mechanism of marine Vibrios [J]. Occupation and Health, 2019, 35(7): 984-989. (in Chinese).
[4] YUAN SB, ZHU AY. Progress on pathogenicity research on Vibrio alginolyticus to aquatic products[J]. Journal of Zhejiang Ocean University: Natural Science Edition, 2012, 31(3): 256-264. (in Chinese).
[5] LU PP, GUO SL, GUAN RZ, et al. Review on the immunogenicity of pathogenic Vibrio outer membrane proteins[J]. Biotechnology Bulletin, 2014(4): 30-35. (in Chinese).
[6] ZUO FQ, JIAN JC, WU ZH. Characterization of extracellular products from Vibrio alginolyticus isolated from maricultured fish[J]. Acta Hydrobiologica SInica, 2006(5): 553-558. (in Chinese).
[7] WANG PB, MA Y, LIU Q, et al. Growth, siderophore production and outer membrane protein expression of Vibrio alginolyticus by iron regulation[J]. Microbiology, 2006(2): 48-53. (in Chinese).
[8] BO KX, ZHOU CX, LU FP, et al. Research on the regulation mechanism of bacterial transcription initiation[J]. China Biotechnology, 2021, 41(11): 89-99. (in Chinese).
[9] ZHANG HZ, KAN B. Progress in study of transcription regulation of cAMP receptor protein[J]. Letters in Biotechnology, 2009, 20(1): 94-98. (in Chinese).
[10] GRAINGER DC, HURD D, HARRISON M, et al. Studies of the distribution of Escherichia coli cAMP-receptor protein and RNA polymerase along the E. coli chromosome[J]. Proceedings of the National Academy of Sciences of the United States of America, 2005, 102(49): 17693-17698.
[11] CHEN SB, ZHANG CJ, CHEN HM, et al. Determination of biological characteristics of Salmonella typhimurium SL1344△crp strain without crp gene[J]. Chinese Veterinary Science, 2015, 45(7): 729-733. (in Chinese).
[12] WICKSTRUM JR, SKREDENSKE JM, KOLIN A, et al. Transcription activation by the DNA-binding domain of the AraC family protein RhaS in the absence of its effector-binding domain[J]. Journal of bacteriology, 2007, 189(14): 4984-4993.
[13] TAE-WOOK NAM, YOUNG-HA PARK, HYE-JIN JEONG, et al. Glucose repression of the Escherichia coli sdhCDAB operon, revisited: Regulation by the CRP center dot cAMP complex [J]. Nucleic Acids Research, 2005, 33(21): 6712-6722.
[14] WEILI L, ALBERTO P-M, J S A, et al. The cyclic AMP receptor protein modulates quorum sensing, motility and multiple genes that affect intestinal colonization in Vibrio cholerae [J]. Microbiology (Reading, England), 2007, 153(Pt 9): 2964-2975.
[15] MANNEH-ROUSSEL J, HAYCOCKS JRJ, MAGN A, et al. cAMP receptor protein controls Vibrio cholerae gene expression in response to host colonization[J]. mBio, 2018, 9(4): e00966-18.
[16] SHANE P, M Y G. Essential role for cyclic AMP and its receptor protein in Yersinia enterocolitica virulence[J]. Infection and immunity, 2002, 70(7): 3665-3672.
[17] LI J, CHEN SB, YU ZH, et al. Construction of host-vector balanced lethal system of Salmonella typhimurium SL1344△cya mutant and immune protection test of chickling[J]. Acta Microbiologica Sinica, 2015, 55(7): 942-948. (in Chinese).
[18] WU PW. Functional study of the C-ring component VscQ of Vibrio alginolyticus T3SS[D]. Zhanjiang: Guangdong Ocean University, 2020. (in Chinese).
[19] STIRLING DA, HULTON CSJ, WADDELL S, et al. Molecular characterization of the proU loci of Salmonella typhimurium and Escherichia coli encoding osmoregulated glycine betaine transport systems[J]. Molecular Microbiology, 1989, 3(8): 1025-1038.
[20] MUKHERJEE S, KEITANY G, LI Y, et al. Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation[J]. Science, 2006, 312(5777): 1211-1214.
[21] ZHAN LJ, YANG L, ZHOU DS, et al. Preliminary investigation on regulation characteristics of virulence-related gene yopD by cAMP receptor protein (CRP) in Yersinia pestis [J]. Chinese Journal of Comparative Medicine, 2009, 19(8): 5-8. (in Chinese).
[22] TAN B. Virulence phenotype analysis of Klebsiella pneumoniae mutant strain Δcrp and regulation of gene KP1_4563 by the regulator CRP [D]. Chongqing: Chongqing Medical University, 2015. (in Chinese).
[23] ZHANG W, CHEN L, LIN L, et al. Molecular cloning, bioinformatics analysis and transcriptional expression of virulence-related gene ( exsA ) of Vibrio alginolyticus [J]. Asian Agricultural Research, 2021, 13(1): 38-42.
[24] PANG HY, ZHOU ZJ, DING Y, et al. Molecular cloning and bioinformatics analysis of T3SS chaperone escort protein VscO from Vibrio alginolyticus [J]. Biotechnology Bulletin, 2014(6): 155-161. (in Chinese).
[25] WU NN, KANG C, RONG N, et al. Bioinformatics analysis of Vibrio alginolyticus TolB protein [J]. Journal of Henan Agricultural Sciences, 2018, 47(11): 134-141. (in Chinese).
[26] LOU H, ZHANG ZW, CHEN QW, et al. Cloning, sequencing and prediction of B cell epitope of structural protein P1 gene of foot-and-mouth disease virus strain A/WH/09 [J]. Veterinary Science in China, 2011, 41(03): 221-228. (in Chinese).
[27] Petzke M M, Suri P K, Bungiro R, et al. Schistosoma mansoni gene GP22 encodes the tegumental antigen sm25: (1) antibodies to a predicted B-cell epitope of Sm25 cross-react with other candidate vaccine worm antigens; (2) characterization of a recombinant product containing tandem-repeats of this peptide as a vaccine [J]. Parasite immunology, 2000, 22(8): 381-395.
[28] DAI JL. Study on the regulatory mechanism of protein phosphorylation modification and toxin-antitoxin system in response to stress in Deinococcus radiodurans [D]. Hangzhou: Zhejiang University, 2021. (in Chinese).
Editor: Yingzhi GUANG Proofreader: Xinxiu ZHU
- 农业生物技术(英文版)的其它文章
- Breeding Technology of Sunflower Inbred Lines with Four Generations in One Year
- Artificial Plant Seeds and Their Application
- Breeding of a Water-saving Drought-resistant Two-line Hybrid Rice Variety Wanliangyou 1008
- Development of Landscaping and Selection of Contemporary Landscaping Plants
- The Register and Characteristics of Chinese Key Protected Wild Plants in Dabie Mountains National Nature Preserve, Hubei Province
- Effects of Uniconazole on Stem and Leaf Growth and Endogenous Hormone Contents of Dahlia ( Dahlia pinnata Cav.)

