N4O2⁃Donor Macrocyclic Schiff Base Ni(Ⅱ) Complex:Synthesis,Crystal Structure,DFT Study and Urease Inhibition Study
WANG HuLOU Qing⁃LiHAO Chun⁃ZhiZHU Wen⁃Xiang YANG YanZHANG WeiMOU DanZHANG XiaYIN Chao⁃Chuang
(1School of Chemistry and Materials Engineering,Liupanshui Normal University,Liupanshui,Guizhou 553004,China)
(2Key Laboratory of Marine Chemistry Theory and Technology,Ministry of Education,College of Chemistry and Chemical Engineering,Ocean University of China,Qingdao,Shandong 266000,China)
Abstract:A Ni(Ⅱ) complex,[Ni(C40H44N4O8)]Cl2·4CH3OH,with a 28⁃membered macrocyclic Schiff base ligand of an N4O2donor set has been prepared by the[2+2]reaction of 1,2⁃bis(2⁃methoxy⁃6⁃formylphenoxy)ethane with ethylene⁃diamine in a 1∶1 stoichiometric ratio in the presence of Ni(Ⅱ) ion employing the template strategy.The complex was characterized by elemental analysis,infrared spectroscopy,powder X⁃ray diffraction,single⁃crystal X⁃ray diffrac⁃tion,and density functional theory quantification calculations.The structural analysis demonstrates that Ni(Ⅱ)ion is six⁃coordinate by four N atoms from C=N groups,two O atoms from ether groups,forming a distorted octahedral geometry.Urease inhibitory activity and molecular docking simulation studies show that the Ni(Ⅱ)complex proved to be a good candidate for the inhibition of jack bean urease.CCDC:1052326.
Keywords:N4O2⁃donor macrocycle;Ni(Ⅱ) complex;crystal structure;DFT calculation study;urease inhibitor
0 Introduction
In the 1960s,synthetic methods for the prepara⁃tion of a range of polyaza,polythia,and polyazathia macrocycles were developed largely employing tem⁃plate strategies that lead to a burgeoning of interest in the field resulting in Schiff bases becoming one of the cornerstones of the field of macrocyclic chemistry[1⁃3].Over the years,NxOymacrocycles especially incorporat⁃ing pyridyl fragments have lent themselves to a range of biological applications which include host⁃guest compounds[4],biomimetics[5],antibiotics[6],as well as metal chelation and extraction[7].In addition to these more commonly reviewed properties,they have also been extensively studied for their magnetic properties to develop new families of probes for medical diagnos⁃tics and potential information storage applications in the form of spin⁃crossover compounds,single⁃molecule magnets(SMMs),and single⁃chain magnets(SCMs)[8⁃10].Thus,considerable attention is given to the preparation and characterization of functionalized NxOymacrocy⁃clic Schiff base complexes[11⁃13].A large number of mac⁃rocyclic multidentate ligand complexes have been investigated extensively,to understand their synthetic,thermodynamic,and structural properties[14⁃19].
So far,most of NxOymacrocyclic Schiff base com⁃plexes mainly refer to the compounds synthesized by reacting 2,6 ⁃diacetylpyridine with a variety ofα,ω⁃diamines in the presence of transition metals or lantha⁃nide ions employing a metaltemplate strategy(Fig.1)[20⁃21].To the best of our knowledge,there is no report on the investigation on the synthesis of 28⁃membered N4O2⁃donor type macrocyclic Schiff base Ni(Ⅱ)complex,its crystal structure analysis,and urease inhibition study using the complex.Given these obser⁃vations,in this work,a new Ni(Ⅱ)complex with a novel N4O2⁃donor macrocyclic Schiff base ligand,namely,[Ni(C40H44N4O8)]Cl2·4CH3OH,has been synthesized,characterized,and evaluated in the urease inhibitory activity.Additionally,based on the crystal data,densi⁃ty functional theory(DFT)study of the complex was performed using the Gaussian 09 program suite.The NBO atomic charges distribution,molecular total ener⁃gy,frontier molecular orbital energies,and molecular electrostatic potential of[Ni(C40H44N4O8)]2+were dis⁃cussed.Moreover,due to the interest in understanding its role in urease inhibition,docking simulation was carried out using the AutoDock Vina program to posi⁃tion the complex into the active site of urease to deter⁃mine the probable binding mode.The docking result showed a good correlation with experimental data.
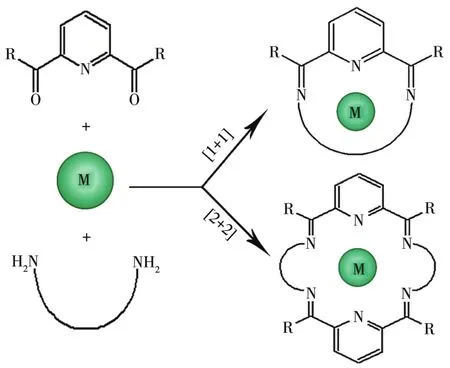
Fig.1 Metal templated synthesis of[1+1]and[2+2]Schiff base macrocyclic complexes
1 Experimental
1.1 Materials and methods
1,2⁃Bis(2⁃methoxy⁃6⁃formylphenoxy)ethane was prepared according to the method given by Tuncer[22].All other chemical reagents used in this work were of analytical grade and were used without further purifica⁃tion.Ethylenediamine was purchased from Aladdin.Urease(from jack beans,typeⅢ,activity:31660 units per mg of solid),N⁃(2⁃hydroxyethyl)piperazine⁃N′⁃(2⁃ethanesulfonic acid)(HEPES)buffer,and urea(molecu⁃lar biology reagent)were purchased from Sigma⁃Aldrich Co.(St.Louis,MO,USA).The infrared spectrum was recorded as KBr pellets on a Nicolet 170SX spectro⁃photometer in the 4 000 ⁃400 cm−1region.Elemental analysis(C,H,and N)was performed in a model 2400 PerkinElmer analyzer.Powder X⁃ray diffraction(PXRD)patterns were obtained on a Shimadzu XRD⁃6000 X⁃ray diffractometer with CuKα(λ=0.154 18 nm)radiation at room temperature.The working volt⁃age was 40 kV,the working current was 40 mA,and the scanning range was 5°⁃40°.Enzyme inhibitory activity was measured with a BioTek Synergy HT microplate reader.
1.2 Synthesis of the Ni(Ⅱ)complex
The general synthetic route of the complex is shown in Scheme 1.
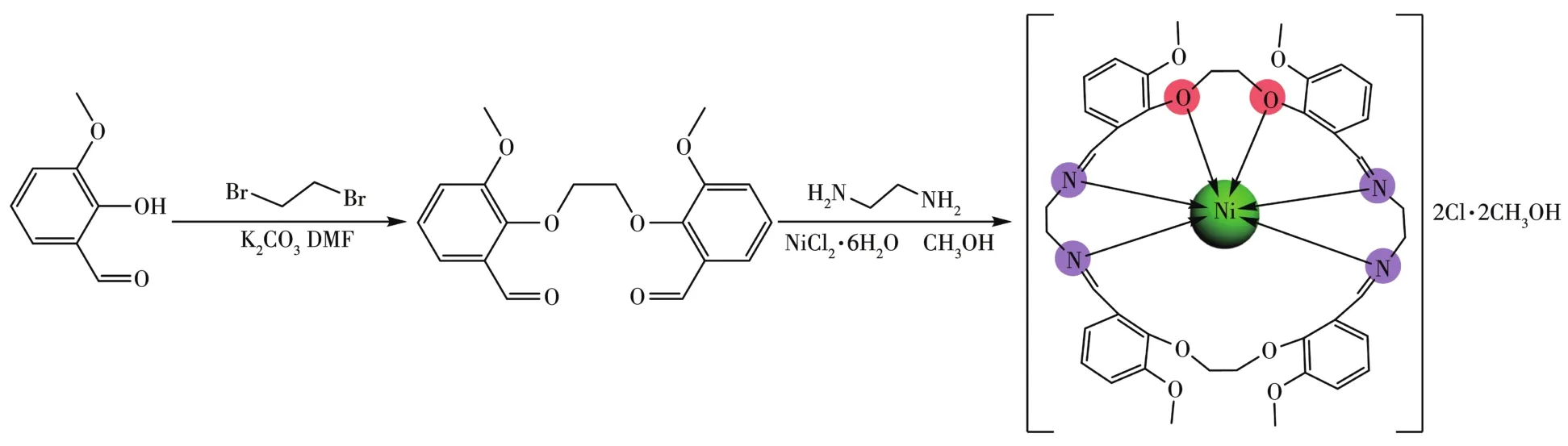
Scheme 1 General synthetic route of the Ni(Ⅱ)complex
1,2⁃Bis(2⁃methoxy⁃6⁃formylphenoxy)ethane(0.330 g,1 mmol)was dissolved in 20 mL of methanol with magnetic stirring and then NiCl2·6H2O(0.119 g,0.5 mmol)was added to the solution.After that,20 mL of methanol solution of ethylenediamine(0.060 g,1 mmol)was dropwise added to the above solution.Then the mixture was stirred and refluxed at 65℃for 4 h.The resulting solution was cooled at room temperature and then filtered.Red columnar⁃shaped crystals were formed after about three days by slow diffusion of ether into a concentrated methanol solution of the complex at room temperature.Yield:63% based on 1,2⁃bis(2⁃methoxy⁃6⁃formylphenoxy)ethane.Elemental Anal.Calcd.for[Ni(C40H44N4O8)]Cl2·4CH3OH(%):C 54.68;H 6.26;N 5.80;Found(%):C 54.71;H 6.23:N 5.82%.IR(KBr,cm−1):1 665s,1 256m,483m,426m.
1.3 Crystallographic data collection and structure determination
For the complex,the single crystal with dimension of 0.48 mm×0.45 mm×0.44 mm was mounted on an Enraf⁃Nonius CAD⁃4 X⁃ray single⁃crystal diffractome⁃ter.All data were collected at 293(2)K with graphite monochromated MoKαradiation(λ=0.071 073 nm)inω⁃2θscan mode.The structure was solved by direct methods using SHELXS⁃97.Non⁃hydrogen atoms were defined by the Fourier synthesis method.Positional and thermal parameters were refined by full⁃matrix least⁃squares(onF2)to convergence.
CCDC:1052326.
1.4 Computational procedure
Becke′s three ⁃parameter hybrid(B3LYP)level was selected for DFT calculations by the basis set of 6⁃311G**for the C,H,N,and O atoms,while valence double⁃ξLANL2DZ basis set was used for Ni atom.Atom coordinates used in the calculations were from crystallographic data,and a molecule in the unit cells was selected as the initial model.All calculations were conducted using Gaussian 09 program.
1.5 Measurement of jack bean urease inhibitory activity
The measurement of urease activity was carried out according to the method reported by Tanaka[23].Generally,the assay mixture,containing 25 μL of jack bean urease(4×104U·L⁃1)(dissolved in distilled water)and 25 μL of the tested complexes dissolved in DMSO/H2O(1∶1,V/V)with different concentrations,was prein⁃cubated for 1 h at 37 ℃ in a 96⁃well assay plate.After preincubation,200 μL of 100 mmol HEPES buffer(pH=6.8)containing 500 mmol urea and 0.002% phenol red were added and incubated at 37℃[24].The reaction was measured by a microplate reader(570 nm),which was required to produce enough ammonium carbonate to raise the pH of a HEPES buffer from 6.8 to 7.7,the endpoint being determined by the color of the phenol red indicator[25].
1.6 Docking simulation
Molecular docking of the inhibitor with the 3D structure of jack bean urease was carried out using the Auto Dock Vina program[26].The ligand structure in docking protocol was used as a crystal structure.The crystal structure of jack bean urease was obtained from Protein Data Bank(http://www.pdb.org/pdb/home/home.do)with the PDB ID of 3LA4S at a resolution of 0.290 nm.The graphical user interface Auto Dock Tools(ADT 1.5.4)was performed to set up every inhibitor enzyme interaction where all hydrogen atoms were added,Gasteiger charges were calculated and nonpolar hydrogen atoms were merged to carbon atoms.A grid box with a size of 6 nm was centered on the active site of the protein.The exhaustiveness parameter was set to 10(default 8).The flexible ligand mode was used for docking.The docking procedure of the Ni(Ⅱ)complex with the active site of jack bean urease was performed as described.
2 Results and discussion
2.1 Structural description of[Ni(C40H44N4O8)]Cl2·4CH3OH
The crystallographic structural analysis reveals that the Ni(Ⅱ)complex crystallizes in the monoclinic crystal system,space groupC2/c(Table 1).Each metal ion is coordinated with six atoms of the macrocyclic Schiff⁃base ligand,namely,four N atoms from C=N groups,two O atoms from ether groups,forming a type of N4O2⁃donor 28⁃membered macrocyclic Schiff base Ni(Ⅱ)complex.

Table 1 Crystallographic data and X⁃ray experiment details for the Ni(Ⅱ)complex
The crystal structure of the Ni(Ⅱ)complex and the coordination geometry of the Ni(Ⅱ)ion of the complex are shown in Fig.2.The selected bond lengths and angles are listed in Table 2.The bond angles of N2#1—Ni1—N2(92.0(2)°)and O1#1—Ni1—O1(78.66°)are both less than 180°,and the bond angles of N1—Ni1—O1(85.56°),N1—Ni1—O1#1(90.98°),N1—Ni1—N2(82.82°),and N1—Ni1—N2#1(100.33°)indicate that Ni(Ⅱ)ion adopts a distorted octahedral geometry(Fig.2,Table 2).Atoms O1,O1#1,N2,and N2#1 occupy each vertex of the basal site,while N1 and N1#1 locate in the apical positions of the octahedral structure.The bond lengths of C1—N1,C1#1—N1#1,C9—N2,and C9#1—N2#1 are 0.126 3(6),0.126 3(6),0.126 4(6),and 0.126 4(6)nm,respectively,which are close to the usual length of C=N[27⁃28],obviously indicating that there are four C=N bonds in the complex:one isbetween C1 and N1,one is between C1#1 and N1#1,one is between C9 and N2,the last one is between C9#1 and N2#1.Furthermore,each ligand serves as a bridging ligand to connect two Ni(Ⅱ) centers into supra⁃molecular structure and the 2D supramolecular layer structure of the complex is constructed by C—O…Cl interactions and C—O…O interactions(Fig.3).
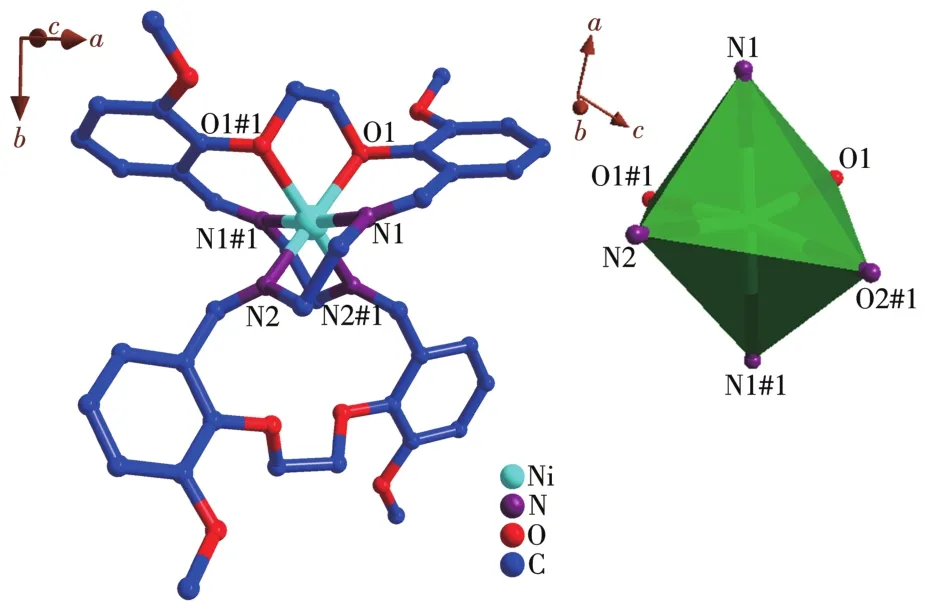
Fig.2 (a)Crystal structure of[Ni(C40H44N4O8)]Cl2·4CH3OH;(b)Coordination geometry of the Ni(Ⅱ)ion of the complex

Table 2 Selected bond lengths(nm)and angles(°)for the Ni(Ⅱ)complex
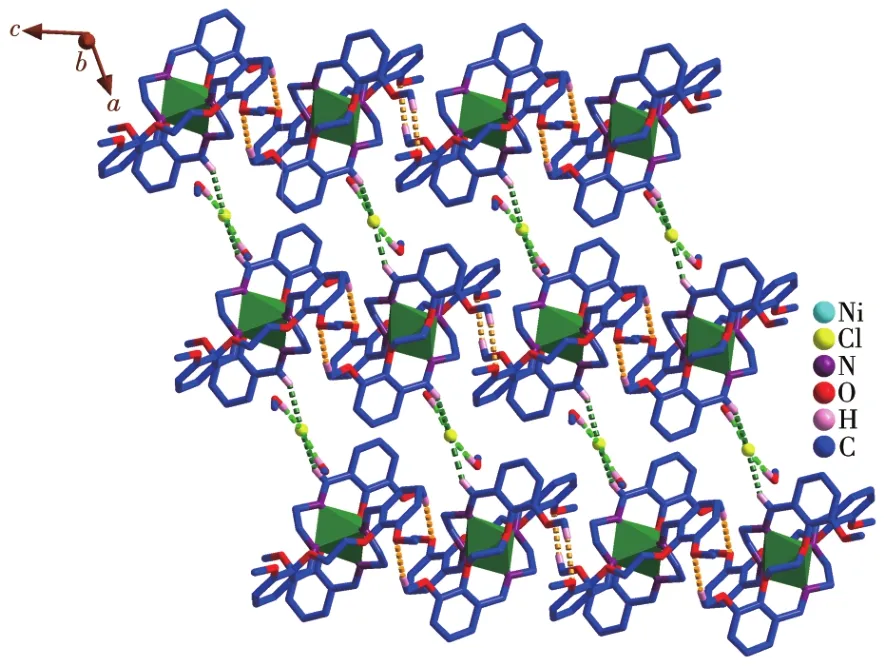
Fig.3 2D layer structure of the Ni(Ⅱ)complex constructed by C—H…Cl interactions(green)and C—H…O interactions(yellow)
2.2 PXRD analysis
The comparison of measured values with the simu⁃lated PXRD patterns is shown in Fig.4.It can be observed that the main characteristic peaks in the two diffraction patterns are identical,which indicates the good phase purity of the complex.The different intensi⁃ty may be derived from the variation in the preferred orientation of the powder samples during the collection of data[29].
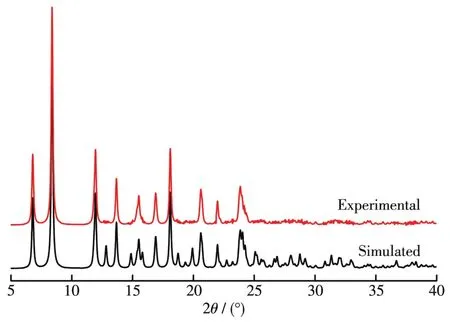
Fig.4 PXRD patterns of the Ni(Ⅱ)complex
2.3 NBO atomic charges and electron configuration
The selected NBO atomic charges and electron configuration for[Ni(C40H44N4O8)]2+are shown in Table 3.It is indicated that the electronic configuration of Ni(Ⅱ)ion,N,and O atoms are 4s0.253d8.64,2s1.34⁃1.392p4.11⁃4.12,and 2s1.60⁃1.622p4.95⁃4.98, respectively.Based on the above results,one can conclude that the Ni(Ⅱ) ion coordina⁃tion with N and O atoms is mainly on 3dand 4sorbit⁃als.Four N atoms form coordination bonds with Ni(Ⅱ)ion using 2sand 2porbitals.Two O atoms supply elec⁃trons of 2sand 2pto Ni(Ⅱ) ion and form the coordina⁃tion bonds.From NBO atomic charges data,we can know that the charge of Ni decreases from 2 to 1.085,which indicates that part of the electrons have trans⁃ferred from the coordinated O atoms and N atoms to the metal ion,and the stable coordination bonds have formed.The negative charges mostly concentrate onthe coordinated O atoms and N atoms,so they are prone to coordinate with Ni(Ⅱ) cation.The results indi⁃cate that the calculation is in accordance with the de⁃termination.
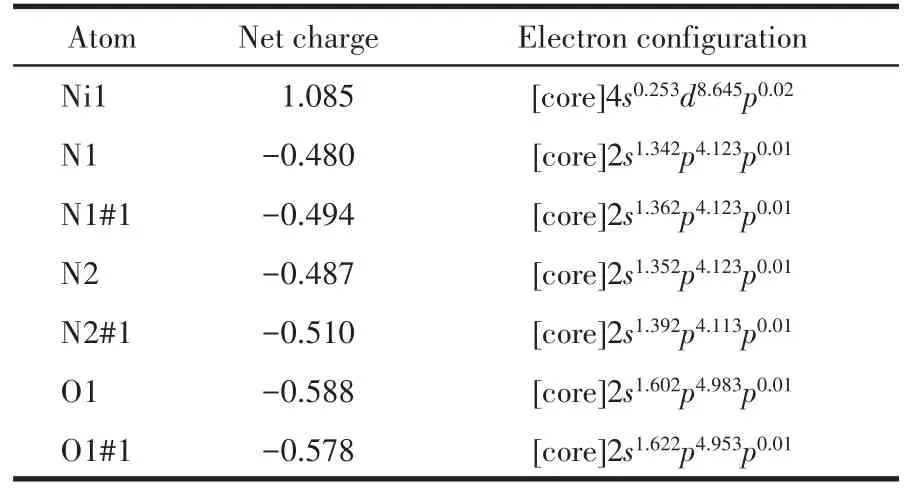
Table 3 Selected NBO atomic charges and electron configuration for[Ni(C40H44N4O8)]2+

Table 4 Inhibition of jack bean urease by the Ni(Ⅱ)complex
2.4 Frontier molecular orbital(FMO)
FMO and its properties such as energy are useful for predicting the most reactive position inπ⁃electron and conjugated systems[30].The energies of HOMO and LUMO and their energy gap reflect the biological activ⁃ity of a molecule[31⁃32].
For the title compound,the energies of HOMO and LUMO are −10.226 and −7.512 eV,respectively.The energies of HOMO and LUMO are negative,which indicates that the studied compound is stable[33].The global chemical descriptors associated with a molecule are:ionization potentialI=−EHOMO=10.226 eV;electron affinityA=−ELUMO=7.512 eV;electronegativityχ=(I+A)/2=8.869 eV;global hardnessη=(I−A)/2=1.357 eV;chemical potentialμ=−(I+A)/2=−8.869 eV;electrophi⁃licity indexω=μ2/(2η)=28.983 eV[34].The HOMO ⁃LUMO plots are given in Fig.5 and from this plot,it is clear that HOMO is mainly localized over the phenyl rings away from the coordination center while the LUMO is mainly localized over the NiN4O2coordina⁃tion unit.This shows that there may be a charge trans⁃fer process in the molecular system[35].
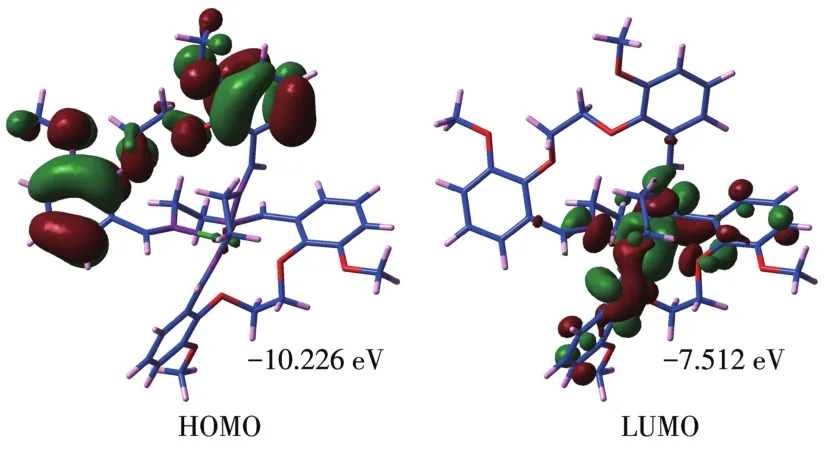
Fig.5 HOMO⁃LUMO plots of[Ni(C40H44N4O8)]2+
2.5 Molecular electrostatic potential(MEP)
MEP is related to electronic density and is a very useful descriptor in understanding sites for electrophil⁃ic attack and nucleophilic reactions.The electrostatic potentialV(r)is also well suited for analyzing processes based on the“recognition”of one molecule by another,as in drug⁃receptor,and enzyme⁃substrate interac⁃tions[36].To predict reactive sites for the electrophilic and nucleophilic process for the studied molecule,the MEP surface was obtained based on the optimized geometry.The negative regions(red)of MEP are relat⁃ed to electrophilic reactivity and the positive(blue)ones to nucleophilic reactivity.
In our present study,the MEP of[Ni(C40H44N4O8)]2+is depicted in Fig.6.The figure shows that there are large positive regions on the whole molecule(blue locality in Fig.6),indicating that there are possible sites for nucleophilic attack.
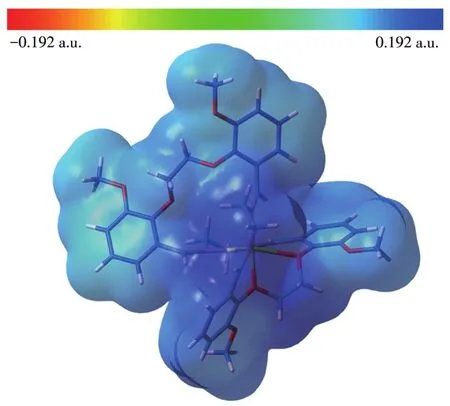
Fig.6 Molecular electrostatic potential map of[Ni(C40H44N4O8)]2+
2.6 Inhibitory activity against jack bean urease
The 1,2⁃bis(2⁃methoxy⁃6⁃formylphenoxy)ethane and the corresponding Ni(Ⅱ)complex were screened for inhibitory activity against jack bean urease.We found that 1,2⁃bis(2⁃methoxy⁃6⁃formylphenoxy)ethane exhib⁃ited no inhibitory ability again the jack bean urease(IC50>100 μmol·L−1).Compared with the standard inhibitor acetohydroxamic acid(AHA,IC50=(26.99±1.43)μmol·L−1),the free Ni(Ⅱ) ion showed a good ure⁃ase inhibitory activity with the IC50value of(20.47±1.58) μmol·L−1.The corresponding IC50value of theNi(Ⅱ)complex as the potential urease inhibitor was(15.36±2.26) μmol·L−1,which was lower than that of the free Ni(Ⅱ) ion.The result indicates that the Ni(Ⅱ)ion coordinated to the Schiff base ligand plays an important role in physiological activity[37⁃38].
2.7 Molecular docking study
The binding model of Ni(Ⅱ)complex with jack bean urease(3LA4)was simulated using the AutoDock Vina program to validate the structure⁃activity relation⁃ship.The result revealed that the target molecule fitted in the active pocket of jack bean urease and gave a binding affinity(ΔG)value of −38 kJ·mol−1(Table 5).The binding model of the title complex with urease(3LA4)is shown in Fig.7a,with all the amino acid resi⁃dues that interact with urease indicated.In the binding model,LYS716 amino acid formed two kinds of cation⁃πinteraction with phenyl rings,with the distances of the interactions being 0.356 4,0.623 2 nm respectively.LYS745 amino acid forms one kind of cation⁃πinterac⁃tion with phenyl ring,with the distance of the interac⁃tion being 0.681 9 nm.
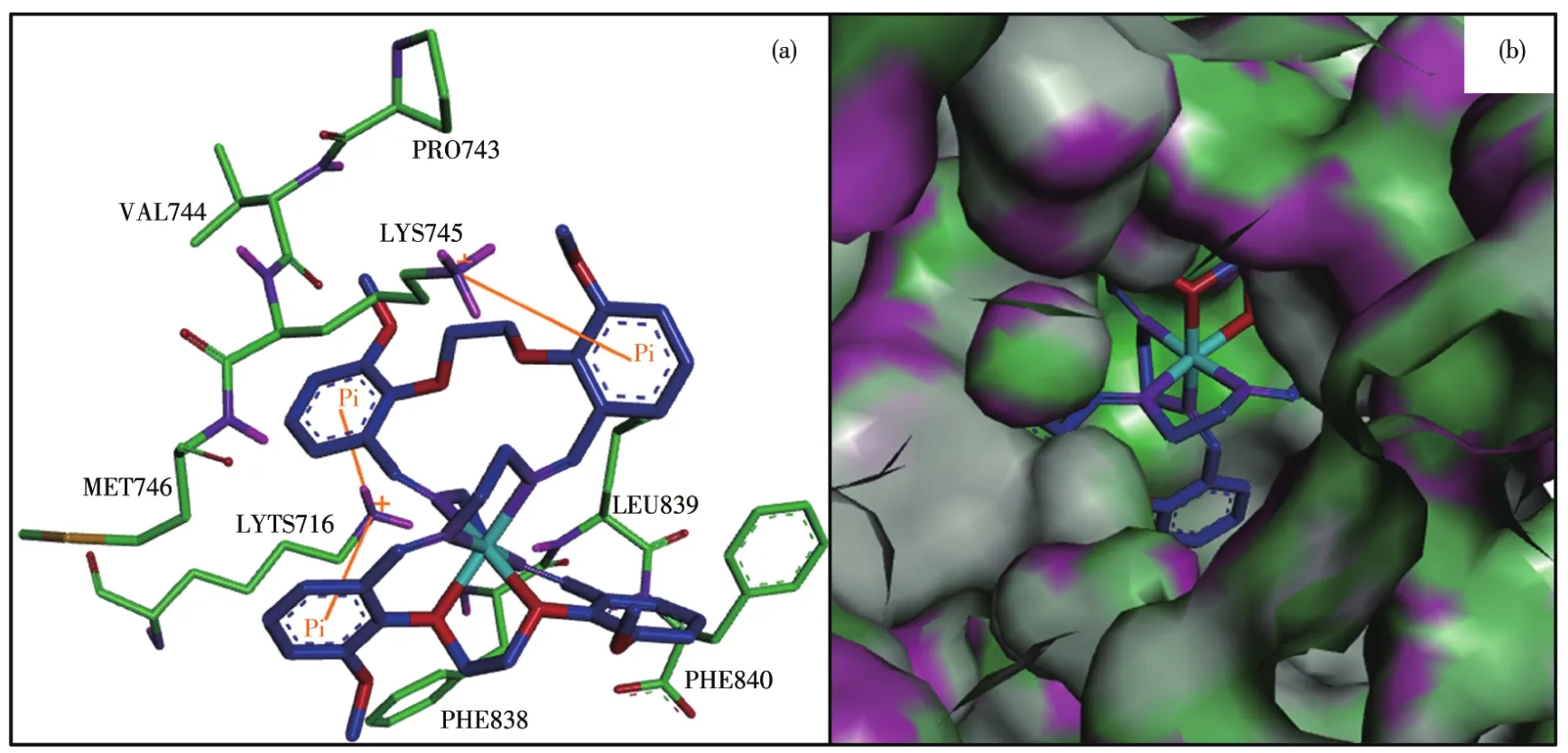
Fig.7 (a)Modeled structure of the urease⁃inhibitor complex,where cation⁃π interactions are presented as yellow lines;(b)Binding mode of the Ni(Ⅱ)complex with jack bean urease where the enzyme is shown as the surface and the complex is shown as sticks
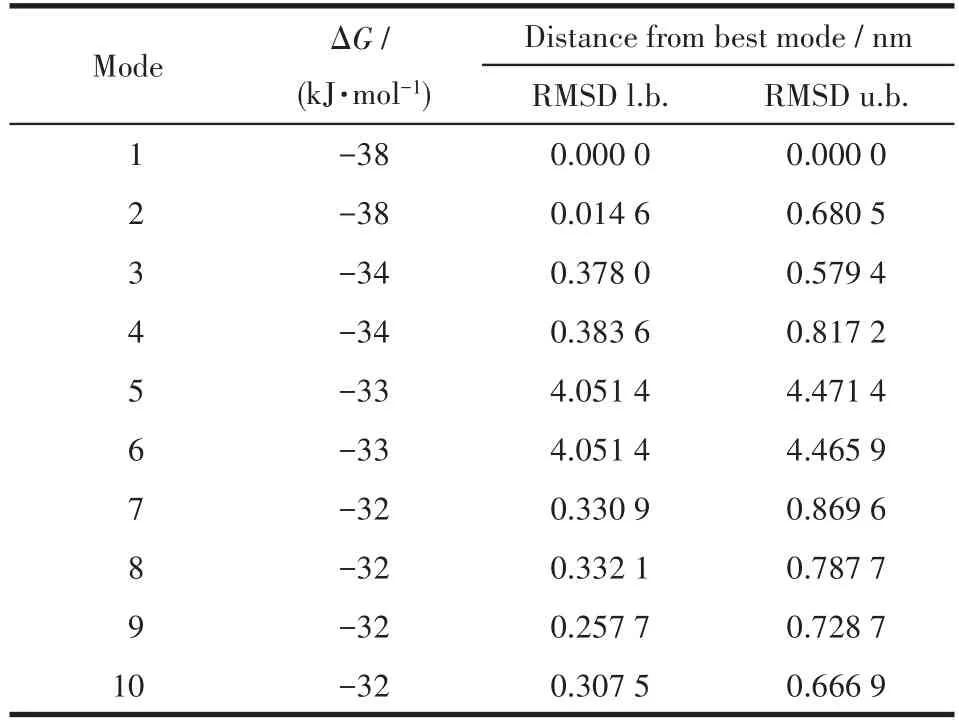
Table 5 Binding affinity values of different poses of the Ni(Ⅱ)complex predicted by AutodockVina
In general,in the inhibitor⁃urease complex confor⁃mation,the Ni(Ⅱ)complex shows a stabilized structure through three types of cation⁃πinteraction between phenyl rings in the complex and the amino acid resi⁃dues.Interestingly,the results of the calculation of the FMO,especially the distribution of HOMO,strongly verify the reasonableness of molecular docking results.This result will intensify our interest in supporting the docking results using the quantum chemistry calcula⁃tion method.
3 Conclusions
In summary,a new Ni(Ⅱ) complex with a 28⁃mem⁃bered N4O2donor macrocyclic Schiff base ligand was synthesized and characterized.Moreover,the inhibitory activity of the Ni(Ⅱ)complex has been testedinvitroagainst jack bean urease and the complex possessed good inhibitory activity.The docking simulation shows that there are three types of cation⁃πstacking interac⁃tions between the complex and the amino acid resi⁃dues.Interestingly,the reasonableness of the docking result was verified by the theoretical study using the DFT method employing a 6⁃311G**basis set.Detailed investigations are continuing to study the mechanism of urease inhibitory activity reported here.
Conflicts of interest:The authors declare no competing financial interest.
Acknowledgments:This work was supported by the Natu⁃ral Science Foundation of Guizhou Province(Grant No.Qiankehe Foundation⁃ZK[2021]General 063),the Youth Talent Growth Project of Educational Department of Guizhou Province(Grants Qianjiaohe KY[2022]No.043,Qianjiaohe KY[2022]No.044),the Scientific and Technological Innovation Platform of Liupans⁃hui(Grant No.52020⁃2021⁃GK⁃04),the Fund of Liupanshui Nor⁃mal University (Grants No.LPSSYKYJJ202002,LPSSYKYJJ202102),and the College Student Innovation Train⁃ing Program(Grant No.202110977033).
- 无机化学学报的其它文章
- 《无机化学学报》投稿须知(NOTICE TO AUTHORS)
- Ag Nanoparticle⁃Doped Activated Microporous Carbon Spheres:An Efficient Adsorption/Catalysis Bifunctional Material for the Removal of Congo Red
- Photocurrent Response Properties of 2,6⁃Bis(3′⁃pyridyl)⁃tetrathiafulvalene Based Zn/Co Coordination Polymer
- A Highly Sensitive and Multi⁃responsive Zn⁃MOF Fluorescent Sensor for Detection of Fe3+,2,4,6⁃Trinitrophenol,and Ornidazole
- Two 3D Microporous Zn⁃MOF for Fluorescence Sensing of Fe3+,Cr2O7 2-,and Acetone in Aqueous Solution
- Structures,Photoluminescent and Magnetic Properties of Three 2D Lanthanide Complexes

