SNHG16 promotes hepatocellular carcinoma development via activating ECM receptor interaction pathway
Qi-Jun Zhng , D-Zhi Li , Bing-Yi Lin , Lei Geng , Zhe Yng , ,, Shu-Seng Zheng ,
a Department of Thyroid Surgery, The First Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou 310 0 03, China
b NHC Key Laboratory of Combined Multi-organ Transplantation, The First Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou 310 0 03,China
c Department of Hepatobiliary and Pancreatic Surgery, Shulan (Hangzhou) Hospital, Zhejiang Shuren University School of Medicine, Hangzhou 310 0 0 0, China
Keywords:Hepatocellular carcinoma SNHG16 ECM-receptor interaction pathway Long non-coding RNA
ABSTRACT
Background: Accumulating data have suggested that long non-coding RNAs (lncRNAs) play important roles in regulating tumor cell growth. This study was designed to investigate the role of SNHG16 in hepatocellular carcinoma (HCC).
Methods: SNHG16 expression was detected with real-time polymerase chain reaction (PCR). The cutoff value of SNHG16 for tumor-free survival (TFS) was determined with receiver operating characteristic curve analysis. Small interfering RNA was used to inhibit the expression of SNHG16 in HCC cell lines.The biologic behavior of HCC cell was determined with cell viability assay and Transwell assay in vitro .The potential predictive value of SNHG16 on prognosis was analyzed by Kaplan-Meier curves and Cox proportional hazards regression model.
Results: SNHG16 expression was upregulated in tumor tissues and HCC cell lines. High expression of SNHG16 was associated with tumor recurrence and poor prognosis after surgery. Multivariate analysis revealed that SNHG16 was an independent prognostic factor for poor recurrence-free survival. Moreover,inhibition of SNHG16 in HepG2, Hep3B, and BEL-7402 cells significantly reduced cell invasiveness and proliferation. Mechanistic analyses indicated that the ECM-receptor interaction pathway was remarkably activated by SNHG16.
Conclusions: SNHG16 might be a promising biomarker for predicting tumor recurrence in HCC patients after surgery and a potential therapeutic target for HCC.
Introduction
Hepatocellular carcinoma (HCC) is the fourth most common cancer and the third common cause of cancer-related deaths worldwide [1]. More than half of the new cases were reported in China due to the high prevalence of hepatitis B virus infection [2] .Although great advance was acquired in HCC therapy, the rate of tumor recurrence and metastasis after surgical resection or liver transplantation (LT) was still high [ 3 , 4 ]. Moreover, the molecular mechanisms underlying tumorigenesis and HCC recurrence were far from understood. Hence, investigation of the essential regulators of HCC would contribute to a deeper understanding of the pathogenesis of the disease and the identification of new therapeutic targets.
Human genomes transcribe abundant RNA molecules. In addition to coding RNAs and short regulatory non-coding RNAs, long non-coding RNAs (lncRNAs) that are longer than 200 nucleotides presented essential functions in regulating cell biology [5] . Recently, emerging studies have implicated that disordered lncRNAs expression are involved in every step of tumor initiation and development including cell proliferation, stemness, migration and invasion, immune escape and drug resistance, etc. [6-8] . Our previous study revealed that suppression of lncRNA00844 expression correlated with poor tumor characteristics, such as portal vein invasion, highα-fetoprotein (AFP), and a high rate of tumor recurrence. Exotic LINC00844 expression inhibits tumor cell proliferation, migration, and invasiveness [9] . Moreover, Notch1 activation specifically induced expression of the lncRNA, TUG1, which regulated self-renewal and stemness of tumor cells [10] . However,the type and molecular mechanism of lncRNAs in HCC remain unclear.
Previously, we examined the transcriptome of eight pairs of HCC tissue samples with RNA sequence, and selected one of the key lncRNAs, SNHG16, as a potential research molecular, which was located on 17q25.1 and significantly increased in HCC. Recent studies have reported that SNHG16 promoted cell growth and invasion, and was correlated with poor prognosis in neuroblastoma and bladder cancer [ 11 , 12 ]. Although SNHG16 has been reported in some solid tumors, its role in HCC is largely unknown. The present study detected the expression of SNHG16 in HCC tissues, and determined its relationship with clinical pathological features and investigated its role in promoting malignancy of HCC cell lines, and analyzed the underlying molecular mechanisms.
Methods
Patients
The present study was conducted according to the principles oftheDeclarationofHelsinki, and approved by the Ethical Committee of the First Affiliated Hospital, Zhejiang University School of Medicine, and informed consent was obtained from all the patients. We included 158 HCC patients undergoing LT at the First Affiliated Hospital, Zhejiang University School of Medicine, and Shulan (Hangzhou) Hospital, Zhejiang Shuren University School of Medicine. Of these patients, HCC was diagnosed by histology.
Cell lines
Normal liver cell line LO2 and ten HCC cell lines (HepG2,Hep3B, SMMC-7721, Huh7, HCCLM3, MHCC-97H, MHCC-97 L, BEL-7402, PLC/PRF/5, and SK-Hep1) were purchased from Shanghai Institute of Cell Biology (Shanghai, China), American Type Culture Collection (Manassas, VA, USA) and Liver Cancer Institute of Fudan University (Shanghai, China). All cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) or 1640 medium supplemented with 10% fetal bovine serum (FBS) in a humidified incubator containing 5% CO2at 37 °C following the standard cell culture techniques.
Quantitative real-time PCR
Total RNA was extracted with TRIzol reagent (Invitrogen, California, CA, USA) from cell lines, tumor tissues and adjacent nontumor tissues. cDNA was synthesized with Takara PrimeScript RT Reagent Kit (Takara Bio, Kyoto, Japan) following the manufacturer’s instructions. The mRNA level of SNHG16 was detected with the Bio-Rad PCR (Thermo Fisher Scientific, Waltham, MA, USA) instrument using SYBR Premix Ex Taq (Takara Bio, Kyoto, Japan). The primer sequences were shown as follows. GAPDH: forward 5 ′ -3 ′ :CCT GGT ATG ACA ACG AAT TTG; reverse 5 ′ -3 ′ : CAG TGA GGG TCT CTC TCT TCC. SNHG16: forward 5 ′ -3 ′ : CAG TCA GCC TCA GTT TCC AA; reverse 5 ′ -3 ′ : AGG CAG GGC TGT GCT GAT.
Cell viability assay
Cell viability was detected with a Cell Counting Kit-8 kit (Dojindo Laboratories, Kumamoto, Japan) according to the manufacturer’s instructions. Briefly, 1 × 103of HCC cells were seeded onto a 96-well plate. After incubating for 24, 48, 72, 96 and 120 h, the culture medium was replaced with fresh medium supplemented with 10 μL CCK-8. The absorbance of cells at 450 nm reflected the cell viability.
Transfection with SNHG16 siRNA
To investigate the role of SNHG16 on HCC function, three HCC cell lines (Hep3B, HepG2, and BEL-7402) were transfected with siRNAs targeting SNHG16 (GenePharma, Shanghai, China) using lipofectamine 20 0 0 transfection reagent (Invitrogen) following the manufacturer’s protocol, and si-GFP was used as a control. The efficacy of SNHG16 siRNA was measured with RT-PCR after transfection for 48 h. The siRNA sequences were shown as follows. si-GFP 5 ′ -3 ′ :CUA CAA CAG CCA CAA CGU CdT dT; si-SNHG16 5 ′ -3 ′ :GGG ACU GCA AGG AUU GUA ATT.
Analysis of cell invasiveness
A 24-well transwell plate (Millipore, Billerica, MA, USA) was used to perform a cell invasion assay. Briefly, 1 × 105HCC cells with si-SNHG16 or si-GFP transfection suspended in 100μL medium without FBS were respectively seeded in the filter coated with Matrigel (BD Bioscience, Franklin, NJ, USA). The lower compartment was filled with 500μL medium supplemented with 10%FBS. After incubating for 48 h, the cells remaining on the upper compartment were removed with cotton swabs, and invasive cells on the bottom surface were stained with 0.4% crystal violet for counting and photographing.
Statistical analysis
All the experiments were repeated three times. The data were presented as mean ± standard deviation or number and percentage. The differences between the two groups were determined by the Chi-square test or two-tailed Student’st-test. The potentially predictive value of SNHG16 on prognosis was analyzed by Kaplan-Meier curves and Cox proportional hazards regression model. All data analysis was conducted with SPSS 19.0 software (SPSS Inc.,Chicago, IL, USA). AP<0.05 was considered statistically significant.
Results
Upregulated SNHG16 expression correlated with poor tumor characteristics in HCC
A total of 118 (75%) HCC tissues showed high SNHG16 expression ( Fig. 1 A). Moreover, SNHG16 expression was higher in HCC cell lines than normal liver cell lines ( Fig. 1 B). The increased expression of SNHG16 was observed in tumors with portal vein tumor thrombus (PVTT) or high AFP value or multiple lesions ( Fig. 1 C-E).
临床药师参与儿童泌尿系统感染的药学会诊实践与分析…………………………………………………… 何翠瑶等(6):852
SNHG16 promotes HCC recurrence following surgery
Receiver operating characteristic (ROC) curve analysis was performed to determine the cutoff value of SNHG16 for tumor-free survival (TFS) and overall survival (OS) [area under the curve (AUC)were 0.656 and 0.681 respectively) and the HCC patients were divided into the high SNHG16 expression group and low SNHG16 expression group ( Fig. 2 A,B). The difference in demographics and tumor characteristics between the two groups was presented in Table 1 . Kaplan-Meier survival analysis showed that 5-year cumulative TFS rate and OS rate were significantly higher in HCC patients with low SNHG16 expression than those with high SNHG16 expression ( Fig. 2 C, D). No matter HCC patients beyond or within the Milan criteria, the SNHG16 kept the predictive significance( Fig. 2 E-H). Besides SNHG16, univariate analysis also disclosed that the largest tumor diameter>5 cm, cumulative tumor size>8 cm,tumor number>3, AFP>400 ng/mL, PVTT, and beyond the Milan criteria were the risk factors for poor prognosis ( Table 2 ). Furthermore, factors withPvalue<0.01 were selected into Cox multivariate analysis model, and further confirmed that cumulative tumor size>8 cm, beyond the Milan criteria, and high SNHG16 expression were independent risk factors for HCC recurrence, and the largest tumor diameter>5 cm, PVTT, and high SNHG16 expression were independent risk factors for OS ( Table 3 ).

Fig. 1. SNHG16 expression was significantly increased in HCC tissues ( A ) and HCC cell lines ( B ) compared with corresponding adjacent normal liver tissues and normal liver cell lines, respectively. Upregulated expression of SNHG16 was correlated with more tumor number ( C ), higher AFP level ( D ), and PVTT ( E ). HCC: hepatocellular carcinoma;AFP: alpha-fetoprotein; PVTT: portal vein tumor thrombus.

Table 1 Patient characteristics.
SNHG16 suppression attenuates HCC cell invasiveness and proliferation
Hep3B, HepG2, BEL7402 with relatively high, medium, and low SNHG16 expression were selected for cell function experimentsinvitro. After siRNA transfection, the SNHG16 expression was apparently suppressed in the above three HCC cell lines ( Fig. 3 A).The invasiveness assay showed that SNHG16-knockdown cell had a significantly decreased invasion capacity compared to negative control cell ( Fig. 3 B). Besides, Hep3B, HepG2, and BEL-7402 with SNHG16-knockdown showed decreased cell viability ( Fig. 3 C).
Verification of SNHG16 function in TCGA database
To further verify the predictive value of SNHG16 in HCC,RNA sequencing data of HCC cases were downloaded from https://xena.ucsc.edu. Transcripts per million (TPM) was used to calculate the relative transcript level. Similar to the above results,SNHG16 expression was significantly upregulated in HCC tissues compared to that in adjacent normal liver tissues ( Fig. 4 A). In this cohort, HCC patients with high SNHG16 expression also showed significantly poorer TFS outcomes ( Fig. 4 B). These results provided solid evidence of SNHG16 as a precise predictor for HCC recurrence after surgery.

Fig. 2. ROC curve analysis was used to determine the cutoff value of SNHG16 for TFS ( A ) and OS ( B ), and the AUC were 0.656 and 0.681, respectively. The 5-year cumulative TFS rate ( C ) and OS rate ( D ) were significantly higher in HCC patients with lower SNHG16 expression than those with increased expression. SNHG16 kept the predictive value for TFS and OS of patients within ( E - F ) or beyond ( G - H ) the Milan criteria. TFS: tumor-free survival; OS: overall survival; HCC: hepatocellular carcinoma; ROC: receiver operating characteristic; AUC: area under the curve.
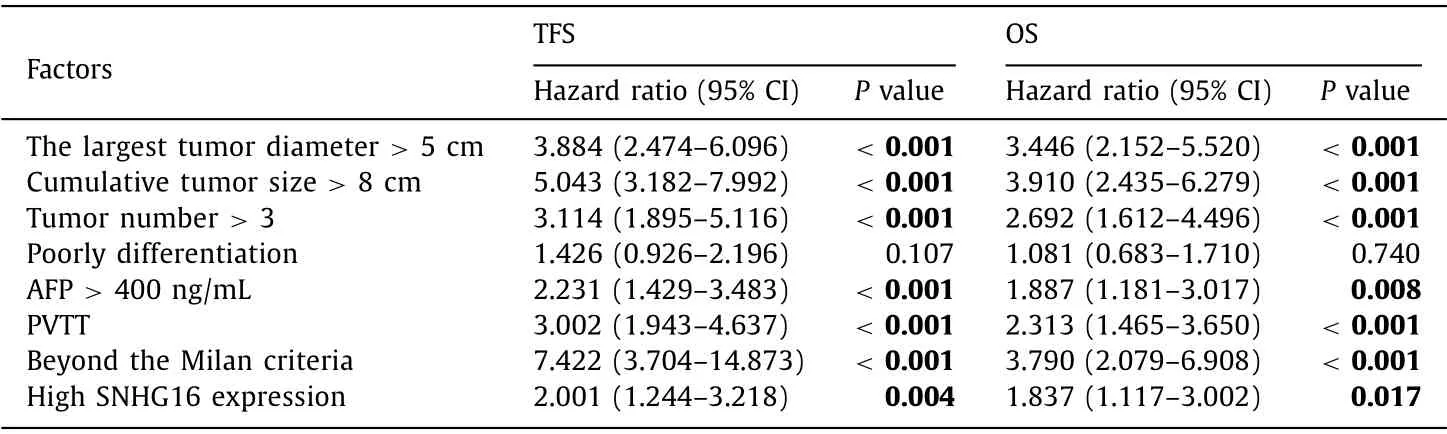
Table 2 Univariate analysis of variables affecting TFS and OS after LT.

Fig. 3. Suppressed expression of SNHG16 in Hep3B, HepG2, and BEL-7402 transfected with siRNA ( A ). Hep3B, HepG2, and BEL-7402 with SNHG16 knockdown showed inhibitory invasive ( B, original magnification × 100), and proliferative ( C ) ability.

Fig. 4. TCGA data confirmed that SNHG16 expression was increased in HCC tissues compared to that in adjacent normal liver tissues ( A ). Patients with high SNHG16 expression showed poorer prognosis following surgery ( B ). HCC: hepatocellular carcinoma; TPM: transcripts per million.
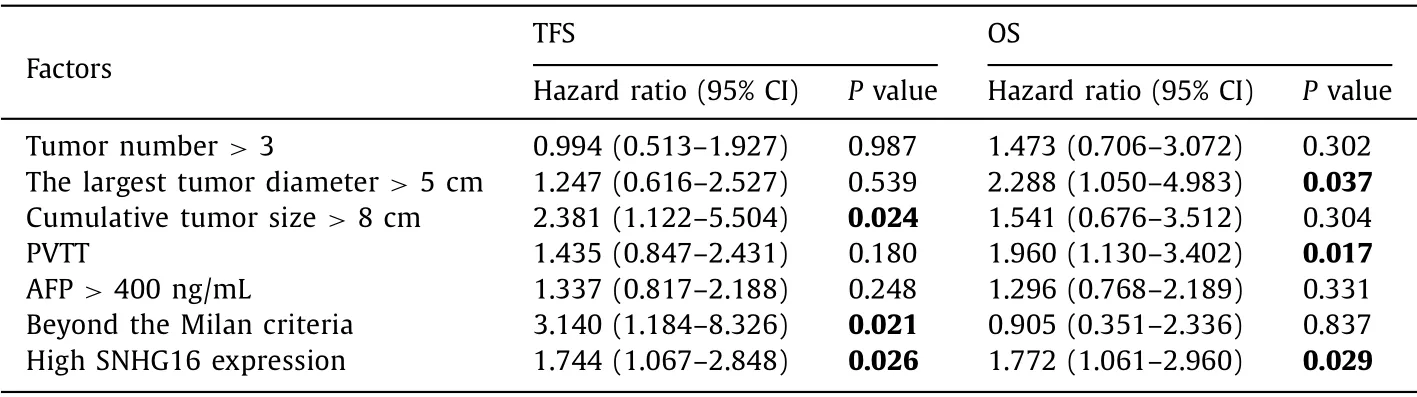
Table 3 Cox proportional hazards regression for predictors of TFS and OS following LT.
SNHG16 involved in ECM-receptor interaction pathways
The gene expression difference analysis was carried out, and a total of 1231 differentially expressed genes were found false positive rate (FDR)<0.05 and fold change (FC)>2 ( Fig. 5 A). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis was utilized to explore the SNHG16 involving in potential signal pathways. The results indicated that SNHG16 regulated four biological pathways including the neuroactive ligandreceptor interaction pathway, the mineral absorption pathway, the ECM-receptor interaction pathway, and the gastric acid secretion pathway ( Fig. 5 B). Among them, ECM-receptor interaction pathway was reported correlating with tumor metastasis and recurrence. Twelve SNHG16 target genes mapped in the ECM-receptor interaction pathway further received Gene Ontology analysis, and demonstrated that they mainly involved in an extracellular matrix organization, collagen-containing extracellular matrix, extracellular region, extracellular space, cell adhesion, and basement membrane( Fig. 5 C). Moreover, of these twelve genes, eleven showed significantly different expression level in HCC tissues compared with adjacent normal liver tissues, indicating that SNHG16 may regulate the mRNA expression of these target genes and affect the ECMreceptor interaction pathway, to promote tumor metastasis and recurrence ( Fig. 6 ).
Discussion
HCC is a highly aggressive and intractable cancer. Although there are advances in preoperative management and surgical techniques, long-term survival outcomes of HCC patients with LT remains challenging due to the high incidence of recurrence and metastasis [13] . Multiple genetic and epigenetic alterations, as well as extrinsic microenvironment factors, contribute to post-surgery HCC recurrence. To date, rare biomarkers could precisely predict the tumor recurrence of HCC and serve as promising therapeutic target [14] . Hence, a better understanding of the molecular mechanism underlying HCC development undoubtedly helps clinical management and improves prognosis. lncRNAs are no longer inconsequential transcription “noise”, which have various molecular biology functions [15] . Recent evidences confirmed that an increasing number of lncRNAs play important roles in human diseases initiation and development. In HCC, a series of lncRNAs with abnormal transcriptional expression due to dysfunctional transcription factors or miRNAs or RNA binding proteins or epigenetically aberrant histone acetylation or DNA methylation were found to be associated with multiple HCC phenotypes [16]. Zhang et al. [17] reported a tumor suppressor lncRNA on chromosome 8p12 termed TSLNC8 that was frequently deleted and downregulated in HCC tissues and suppressed the proliferation and metastasis of HCC cellinvitroandinvivoby inactivating the interleukin-6/STAT3 signaling pathway. Wei et al. [18] reviewed a set of lncRNAs involved in HCC resistance to sorafenib, adriamycin, 5-fluorouracil, and platinum drugs. Moreover, lncRNAs also were reported to promote self-renewal of HCC stem cell through activating the Wnt signaling pathway [19] , and to induce HCC immune evasion by inhibiting the polarization of M2 macrophages [20] . These studies demonstrated that lncRNAs could be considered potential predictors of prognosis and enable the design of precise therapy target for HCC. These findings inspired us to detect the lncRNAs expression profile from eight pairs of HCC tissue samples with RNA sequence to further investigate the HCC related lncRNAs [21] .
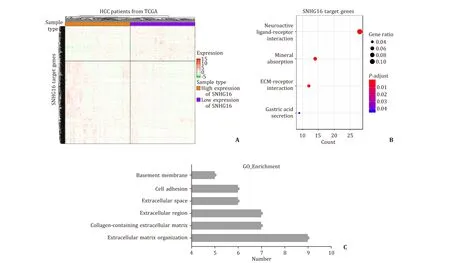
Fig. 5. A total of 1231 genes expressional level were potentially influenced by SNHG16 ( A ). KEGG analysis showed that SNHG16 involves in the mineral absorption pathway,the neuroactive ligand-receptor interaction pathway, the ECM-receptor interaction pathway, the gastric acid secretion pathway ( B ). Gene Ontology analysis demonstrated that SNHG16 correlates with extracellular matrix organization, collagen-containing extracellular matrix and cell adhesion, etc. ( C ). HCC: hepatocellular carcinoma; ECM:extracellular matrix.

Fig. 6. Except for FRAS1 ( C ), the other eleven genes expression were significantly different in HCC compared with adjacent normal liver tissues. T: tumor tissue; N: normal tissue; ECM: extracellular matrix.
Our results confirmed that SNHG16 expression was upregulated in HCC cell lines and HCC tissues compared to that in normal liver cell lines and adjacent normal liver tissues respectively. Furthermore, patients with a low expression level of SNHG16 had a higher TFS rate. Cox proportional hazards regression model analysis showed that SNHG16 was an independent risk factor for HCC recurrence. The predictive value of SNHG16 in HCC was also supported by the TCGA cohort. Recently, many studies have criticized that Milan criteria are too strict for HCC patients in LT waiting list because patients beyond the Milan criteria may also have a low risk of recurrence [ 22 , 23 ]. Therefore, identifying a novel predictor to classify the HCC patients beyond the Milan criteria with a low risk of tumor recurrence, and expand the potential receptors pool is necessary. Our results showed that SNHG16 is a novel and precise biomarker for HCC patients’ prognosis, and may serve as a factor for selecting LT candidates.
In conclusion, SNHG16 expression is significantly increased in HCC tissues and cell lines, correlating with poor prognosis of HCC patients. SNHG16 promotes HCC cell malignant behavior via activating the ECM-receptor interaction pathway.
Acknowledgments
None.
CRediT authorship contribution statement
Qi-Jun Zhang : Formal analysis, Resource, Writing - original draft. Da-Zhi Li : Data curation, Software, Supervision. Bing-Yi Lin :Data curation, Validation, Visualization. Lei Geng : Project administration. Zhe Yang : Conceptualization, Funding acquisition, Investigation, Writing - review & editing. Shu-Seng Zheng : Conceptualization, Supervision.
Funding
The study was supported by grants from Science and Technology Projects of Medicine and Health in Zhejiang Province( 2020383364 ) and Natural Science Foundation of Zhejiang Province( LY21H160055 ).
Ethical approval
The present study was conducted according to the principles oftheDeclarationofHelsinki, and approved by the Ethical Committee of the First Affiliated Hospital, Zhejiang University School of Medicine, and informed consent was obtained from all the patients.
Competing interest
No benefits in any form have been received or will be received from a commercial party related directly or indirectly to the subject of this article.
 Hepatobiliary & Pancreatic Diseases International2022年1期
Hepatobiliary & Pancreatic Diseases International2022年1期
- Hepatobiliary & Pancreatic Diseases International的其它文章
- Targeting pancreatic ductal adenocarcinoma: New therapeutic options for the ongoing battle
- How open is the therapeutic horizon for pancreatic cancer patients?
- Terlipressin versus placebo in living donor liver transplantation
- Fas -670 A/G polymorphism predicts prognosis of hepatocellular carcinoma after curative resection in Chinese Han population
- Meso-Rex bypass for the management of extrahepatic portal vein obstruction in adults (with video)
- The effect of SphK1/S1P signaling pathway on hepatic sinus microcirculation in rats with hepatic ischemia-reperfusion injury
