Isolation and Characterization of A High-efficiency Atrazinedegrading Strain Paenarthrobacter ureafaciens ZF1
Zhang Zhi-fei,Ding Ming-yue,Fu Qian,and Liu Rong-mei
College of Life Sciences,Northeast Agricultural University,Harbin 150030,China
Abstract:The widely use of atrazine causes great threat to the environment.Microbial degradation is an effective method to quickly remove atrazine to maintain environmental safety.A bacterial strain,ZF1,capable of highly degrading atrazine which could grow with atrazine as both carbon and nitrogen sources,was isolated from the wheat field soil in Henan Province,China.Phenotypic characterization and 16S rRNA gene sequencing indicated that the isolate belonged to the Paenarthrobacter ureafaciens.Polymerase chain reaction (PCR) analysis revealed that ZF1 contained the atrazine-degrading genes trzN,atzB and atzC.The growth and atrazinedegradation efficiency of strain ZF1 had broad ranges of temperature (5℃-50℃) and pH (5.0-11.0),and had a high tolerance to atrazine and could tolerate at least 1 500 mg · L-1.The addition of carbon and nitrogen sources promoted the degradation rate of atrazine to a certain extent.When the strain was cultured with starch as carbon sources,the degradation rate of atrazine could reach 50 mg · L-1 · h-1.In general,the discovery of strain ZF1 enriched the resources of atrazine degrading strains.Its excellent biodegradability and environmental tolerance showed that strain ZF1 had great potential in the bioremediation of atrazine contaminated sites in the future.
Key words:atrazine, Paenarthrobacter ureafaciens ZF1,biodegradation
Introduction
Atrazine (2-chloro-4-ethylamino-6-isopropylamino-1,3,5-triazine) is a triazine herbicide with high selective and endothermic properties.In the past 40 years,it has been widely used in the control of broad-leaved weeds,such as corn,wheat,sorghum,sugarcane and annual grasses (Quet al.,2017).However,due to its long half-life,easy diffusion and high mobility,long-term use will cause serious pollution to soil,surface water and groundwater (Liuet al.,2002;Menget al.,2009).In addition,toxicological studies have shown that long-term exposure to atrazine can interfere with human endocrine system,and is prone to carcinogenesis and teratogenesis (Cooperet al.,2000;Luanet al.,2003).Considering the widespread use of atrazine and its toxicity,it is crucial to find a practical solution for the remediation of atrazine-contaminated environments.
Microbial degradation is a cost-effective,environmentally friendly and highly efficient method that has been exploited for remediation of atrazine-contaminated sites (Zhouet al.,2008;Liuet al.,2010;Liet al.,2014;Sagarkaret al.,2014).Currently,many strains that can degrade atrazine have been isolated.These strains includePseudomonas aeruginosa(Fernandeset al.,2014),Arthrobactersp.C2 (Caoet al.,2021),Shewanellasp.YJY4 (Yeet al.,2016),Bacillus subtilisHB-6 (Wanget al.,2014),Acinetobacter lwoffiiDNS32 (Guoet al.,2012) andCitricoccussp.TT3 (Yanget al.,2018).Pseudomonassp.ADP (Lin,2014) andArthrobactersp.TC1 (Stronget al.,2002)are well studied model strains.ADP can degrade atrazine by degrading six enzymes encoded byatzA,atzB,atzC,atzD,atzEandatzF,and finally atrazine can completely mineralize CO2and NH3(Desittiet al.,2016).TC1 contains related degradation genestrzN,atzBandatzC,which encode three enzymes that degrade atrazine into cyanuric acid (Sajjaphanet al.,2004).The atrazine-degrading genesatzAandtrzNuse the same biochemical mechanism,as doatzDandtrzD(Karns,1999;Meyeret al.,2009;Maet al.,2017;Rehanet al.,2017).
To the best of our knowledge,the screening of atrazine degrading strains is mainly concentrated in northeast China (Zhanget al.,2008;Zhanget al.,2012;Li,2017;Liet al.,2019).There are few studies on atrazine degrading strains in other regions.Moreover,the biodegradation efficiencies of these bacteria are not efficient enough in most cases (Zhenget al.,2009;Liet al.,2010;Yanet al.,2011).
Therefore,it is necessary to expand the regional scope of strain screening and isolate new bacterium which has much higher atrazine removal efficiency than common bacteria.In this study,the bacterial strainPaenarthrobacter ureafaciensZF1,which was capable of degrading atrazine,was isolated from the wheat field soil in Henan Province,China.The genes involved in atrazine degradation were investigated.
In addition,the effects of various culture conditions(such as temperature,pH,initial atrazine concentration,carbon and nitrogen sources) on atrazine degradation by the bacterial strain were examined,and the optimum culture conditions for atrazine degradation by strain ZF1 were determined.The work broadened the geographical scope of strain screening and provided a new option for the bioremediation of atrazinecontaminated sites.
Materials and Methods
Sample collection
Soil samples were collected from the wheat field soil in Henan Province,China,in which atrazine had been used for a long time.The topsoil (0-10 cm) was sieved(2.0 mm) to remove stone and plant debris,and then the samples were stored at 4℃ for the further study.
Chemicals and media
Atrazine (purity: 99.8%) and cyanuric acid (purity:98.5%) was purchased from Sinopharm Chemical Reagent Co.,Ltd.(Shanghai,China),methanol used for high-performance liquid chromatography (HPLC)analysis was of HPLC grade,other chemicals used in this study were of analytical grade.
The liquid enrichment medium used was a mineral salts medium (MSM),and each liter of medium containing 0.6 g H2PO4,4.0 g K2HPO4,0.3 g MgSO4· 7H2O,0.6 g NaCl and 3 g glucose.Glucose was used as the carbon source,atrazine with a certain concentration was used as the only nitrogen source for growth.Finally,pH of MSM culture medium was adjusted to 7.2 and sterilized at 115℃ for 20 min.
The LB medium used contained 10 g NaCl,5 g yeast powder and 10 g tryptone per liter.For the solid medium,12 g agar was added to the above liquid medium,and sterilized at 121℃ for 20 min.
Enrichment and isolation of atrazine-degrading bacteria
Five gram soil specimens were added to a 500 mL Erlenmeyer flask containing 100 mL atrazine-MSM(50 mg · L-1atrazine).These specimens were cultured on a 150 r · min-1shaker for 7 days and the temperature was kept at 30℃.Then,5 mL enrichment culture was transferred to fresh AMSM,at this time,the concentration of atrazine increased to 100 mg · L-1.This process was repeated four times until the final atrazine concentration was 200 mg · L-1.Eventually,the medium was diluted and placed onto the agar plates that contained 100 mg · L-1of atrazine,and cultured at 30℃.Different colonies in morphology were selected and cultured to separate pure culture.Single colony of each isolate was transferred to 100 mL AMSM (100 mg · L-1atrazine) to test its biodegradation ability.
ldentification of strain ZF1
The identification of strain ZF1 was based on the conventional physiological and biochemical identification methods,and was confirmed by PCR amplification of 16S rRNA gene using the genomic DNA of strain ZF1 as template.The oligonucleotide primers used were 27F (5'-AGAGTTTGATCCTG GCTCAG-3') as the forward primer and 1492R (5'-GGTTACCTTGTTACGACTT-3') as the reverse primer (Barchanskaet al.,2017).The PCR conditions were as the followings: 94℃ for 5 min followed by 30 cycles of 95℃ or 1 min,54℃ for 1 min and 72℃for 2 min,with a final extension of 10 min at 72℃(Liet al.,2019).The products of PCR were divided on a 1.2% agarose gelelectrophoresis.The resulting DNA fragments were purified through AxyPrep DNA gel extraction kit (Axygen),genetic sequencing was completed by Jilin Comate Bioscience Co.,Ltd.,and the results were compared using the Blast program in the NCBI GenBank nucleotide database.Phylogenetic and molecular evolutionary analyses were conducted using MEGA version 6.0.The tree was constructed using the neighbor-joining method.
Analysis of atrazine-degrading gene
To determine whether strain ZF1 contained these genes,the total genomic DNA was extracted as template and the PCR was performed using previously reported primers (Martinezet al.,2001;Mulbryet al.,2002).The PCR reaction was performed using the following rmocycler program: 5 min at 95℃;30 cycles of 1 min at 95℃,1 min at the optimal temperature for annealing and 2 min at 72℃;then,an additional 10 min cycle at 72℃.The PCR products were detected by electrophoresis using a 1%agarose gel.Sequencing and sequence analyses were conducted as described above.
Degradation and growth of strain ZF1
The MSM culture medium containing atrazine (100 mg · L-1) or cyanuric acid (100 mg · L-1) was used for degradation experiment,among them,atrazine or cyanuric acid was used as nitrogen source and glucose (3 g · L-1) was used as carbon source.Bacterial suspensions (1×108cfu · mL-1) were inoculated at 1%(V/V) of the culture volume,then,incubated at 30℃and 150 r · min-1.During the 6-hour incubation,OD600and the concentration of atrazine and cyanuric acid were measured every half hour to monitor cell growth and atrazine degradation,respectively.Each treatment was carried out in triplicate.
Determination of atrazine and cyanuric acid levels
The enriched culture solution was extracted with two times the volume of dichloromethane,and then,the organic extract was concentrated and evaporated to dryness using a rotary evaporator,dissolving residual atrazine with HPLC grade methanol,aqueous samples were filtrated through a 0.22 μm nylon filter and stored in brown liquid chromatography bottle,and the degradation rate of atrazine was determined by HPLC.The enriched culture solution was directly centrifuged,and the water-containing sample was filtered by 0.22 micron nylon filter for the determination of cyanuric acid.The concentration of atrazine and yield of cyanuric acid from each sample were quantified by HPLC (Agilent 1 260 Infinity ℃).For atrazine detection,the HPLC was fitted with a reverse-phase C18 column (4.6 mm×100 mm,5 μm) with a variablewavelength UV detector set to 215 nm.The flow rate was set to 1.0 mL · min-1(methanol/water=80/20,V/V) and column temperature was set to 30℃.The detection conditions of cyanuric acid were as the followings: the mixture of 0.002 mol · L-1potassium dihydrogen phosphate and 0.005 mol · L-1dipotassium hydrogen phosphate: methanol=95 : 5 (V : V),the column temperature was 35℃,the injection volume and flow rate were the same as above,and the detection wavelength was 213 nm.
Calculation of atrazine degradation rate
According to the results of HPLC determination,the degradation rate of atrazine was calculated,and the percentage of atrazine degradation could be calculated as:
X=(CCK-CX)/CCK×100%
Where,Xwas the degradation rate of atrazine,CXwas the final concentration of atrazine (mg · L-1),andCCKwas the original concentration of atrazine (mg · L-1).
Growth and biodegradation capacity of strain ZF1 and influential factors
In order to determine the influence of external environment (substrate concentration,temperature and pH) on the growth and atrazine degradation ability of the strain,the temperature was adjusted to 5℃,10℃,20℃,30℃,35℃,40℃,45℃ and 50℃,respectively;pH was adjusted to 4.0,5.0,6.0,7.0,8.0,9.0,10.0,11.0 and 12.0,respectively;the concentration was adjusted to 50,100,200,500,750,1 000,1 500 and 2 000 mg · L-1,respectively.Bacterial suspensions (1×108cfu · mL-1) were inoculated into a 100 mL MSM medium with the transfer of 1% (V/V)of the culture volume.Glucose (3 mg · L-1) was the only carbon source and atrazine (100 mg · L-1) was the only nitrogen source in all the treatments.Oscillating culture under different conditions according to experimental requirements,growth of atrazine-degrading bacteria and the concentration of atrazine were measured after 2 and 3.5 h of cultivation.When determining the influence of a single variable,ensured that other culture conditions were the same.These treatment groups were cultured on a shaker at 150 r · min-1,and samples were taken regularly to determine the growth of atrazine-degrading bacteria and the concentration of atrazine.All the experiments were repeated three times.
At the same time,in order to study the effects of different carbon and nitrogen sources on the growth of strains and the degradation of atrazine,in this study,glucose,sucrose,lactose and sodium citrate(3 g · L-1) were used as carbon sources,and ammonium NH4NO3,urea,KNO3and NH4Cl (1 mg · L-1) were used as additional nitrogen sources to explore the effects of exogenous carbon and nitrogen sources on atrazine degradation.Bacterial suspensions (1×108cfu · mL-1)were inoculated into a 100 mL MSM medium with the transfer of 1% (V/V) of the culture volume.The treatments used were as the followings: AT+C:glucose,sucrose,starch or sodium citrate (3 g · L-1) as the carbon source and atrazine (100 mg · L-1) as the sole nitrogen sources;AT+N: NH4NO3,urea,KNO3,NH4CL (1 g · L-1) as the additional nitrogen source and glucose (3 g · L-1) as the sole carbon source.At the same time,a sample without additional nitrogen source was set as a blank control;and AT: no additional carbon or nitrogen sources,atrazine as the sole carbon and nitrogen sources.Incubated all the samples with shaking at 150 r · min-1and 30℃.Samples were taken at 2 and 3.5 h,respectively,and the bacterial concentration and the residual concentration of atrazine in the samples were determined.There were three repetitions for each treatment group.
Statistical analysis
One-way analysis of variance (ANOVA) was used to analyze the significant differences among treatments.Origin 9.0 (Origin Lab,USA) was used for statistical computing and plotting.
Results
lsolation and identification of strain ZF1
A bacterial strain,ZF1,was isolated from the wheat field soil in Henan Province,China.Its physiological and morphological characterizations indicated that ZF1 was a gram-positive,yellow-pigmented and coccoid-shaped bacterium (Fig.1).Its 16S rRNA gene sequence had a size of 1 374 bp.A comparison of the 16S rRNA gene sequence of strain ZF1 with the available sequences in the GenBank database suggested that this strain had 99.56% homology withPaenarthrobacter ureafaciens.Subsequently,a phylogenetic tree of similar 16S rRNA gene sequences was constructed using the neighbor-joining method(Fig.2).In this tree,strain ZF1 was in the same branch asPaenarthobacter ureafaciens.The morphological characteristics of the colony also indicated that this strain belonged toPaenarthrobacter ureafaciens.The sequence of strain ZF1 had been submitted to GenBank with accession number OL415511.

Fig.1 Colony of strain ZF1

Fig.2 Phylogenetic tree based on 16S rRNA gene sequences of Paenarthrobacter ureafaciens ZF1
Analysis of degradation genes of strain ZF1
Using the genome of the strain ZF1 as a template for polymerase chain reaction analysis,432 bptrzN,493 bpatzBand 579 bpatzCwere obtained (Fig.3).Despite the use of various PCR conditions with different primer pairs,the other five genes were not successfully detected.The results of sequence determination and comparison demonstrated that the nucleotide sequences oftrzNandatzBfrom

Fig.3 Agarose gel electrophoresis (1.0%) of PCR products of atrazine-degrading genes of strain ZF1
Paenarthrobacter ureafaciensZF1 showed 99.31%and 100% sequence similarities withArthrobactersp.DNS10 (GenBank accession number KF453507)(Zhanget al.,2011).TheatzCgene was 100.00%homologous withatzCgene ofPaenarthrobacter ureafaciensCS3 (GenBank accession number MF612193) (Yanget al.,2018).The PCR experiment results showed that strain ZF1 had the same degradation genes as many reported atrazine metabolism bacteria,includingtrzN,atzBandatzCgenes,such asArthrobactersp.X-4 (Yanet al.,2011) andSinorhizobiumsp.1128 (Quet al.,2008).These strains could only degrade atrazine to cyanuric acid and it was speculated that strain ZF1 could also degrade atrazine to non-toxic cyanuric acid.
Growth and degradation characteristics of strain ZF1
The growth of strain ZF1 was measured by UV spectrophotometry,and the removal of atrazine was analyzed by HPLC.The results showed when strain ZF1 used cyanuric acid as the sole nitrogen source,the strain hardly grew,indicating that strain ZF1 could not use cyanuric acid as the sole nitrogen source to grow(Fig.4).With atrazine as the sole nitrogen source and glucose as the carbon source,strain ZF1 grew slowly within 1.5 h after inoculation.But its concentration increased significantly after 1.5 h,during this period,the strain ZF1 grew rapidly,and then tended to be flat after 5 h.With the rapid growth of the strain,the concentration of atrazine in the medium decreased rapidly (Fig.5),and completely removed atrazine(100 mg · L-1) within 3.5 h.This indicated that the strain had strong adaptability,could grow rapidly in a short period of time,and quickly use atrazine as a nitrogen source to achieve the effect of rapid degradation of atrazine.The degradation of atrazine was significantly positively correlated with strain growth.Cyanuric acid concentrations detected using HPLC increased gradually over time to a maximum of about 68.38 mg · L-1(Fig.5).This indicated that strain ZF1 might be able to transform atrazine only to cyanuric acid,but be unable to fully mineralize atrazine.This was consistent with the results of the PCR molecular assay.
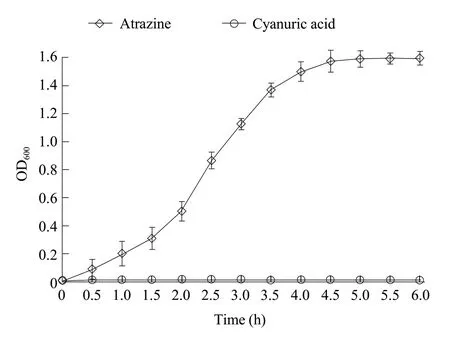
Fig.4 Growth curve of strain ZF1 in MSM medium containing either atrazine or cyanuric acid (100 mg · L-1) as the sole nitrogen source

Fig.5 Growth of strain ZF1 and degradation of atrazine by strain ZF1
Effects of environmental factors on growth and biodegradation
The strain ZF1 was cultured for 2 and 3.5 h under the conditions of different temperatures,pH,substrate concentrations and additional carbon or nitrogen sources.The growth of the strain and the degradation of atrazine are shown in Fig.6.The optimal temperature range for the growth and degradation of atrazine by strain ZF1 was 30℃-40℃ (Fig.6a).When the temperature was 35℃,the degradation rate of atrazine (100 mg · L-1) reached 93.87% within 2 h.When the temperature decreased to 5℃ or increased to 50℃,the degradation efficiency of strain ZF1 atrazine still reached 49.93% and 63.19%,respectively within 2 h and the growth of strain ZF1 also maintained a good state.This showed that strain ZF1 had strong cold and heat resistance,and could adapt to the ecological environment of cold and high temperature.

Fig.6 Growth and atrazine degradation by strain ZF1 under different culture conditions
Fig.6b showed the biodegradation of atrazine by strain ZF1 incubated for 2 and 3.5 h at different pH levels.When pH was 7.0,the degradation rate of atrazine and the growth of strain ZF1 were the greatest.The degradation rate of atrazine gradually decreased as pH increased or decreased.When pH was higher than 11.0 or lower than 5.0,it had a great influence on the degradation ability and growth of the strain.But it had a relatively good degradation effect at pH 6.0-11.0,demonstrating that pH range for atrazine biodegradation by ZF1 was wide and alkaliphilic.
Fig.6c showed that when the concentration of atrazine was lower than 750 mg · L-1,the strain ZF1 could completely degrade atrazine within 3.5 h.When the concentration of atrazine increased by 1 000 mg · L-1,the degradation rate of atrazine was 98.07% in 3.5 h,indicating that strain ZF1 could tolerate higher concentration of atrazine.The OD600of strain ZF1 increased with the increase of atrazine concentration,which might because high concentration of atrazine provided more abundant nitrogen sources and energy (Yanget al.,2018).However,when the concentration of atrazine increased to 1 500 mg · L-1,the degradation rate of atrazine was 97.72%,but the concentration of bacteria ZF1 decreased within 3.5 h,indicating that high atrazine concentrations might inhibit the growth of strain ZF1 (Liet al.,2018).The OD600was not measured,when the concentration of atrazine was higher than 2 000 mg · L-1,because atrazine had low water solubility and thus could not be completely dissolved in the culture medium.
During the whole culture process,atrazine (100 mg · L-1)in the treatment group with starch as carbon source was completely degraded after 2 h of culture (Fig.6d).The bacterial concentration of strain ZF1 was also higher than that of other treatment groups,indicating that starch was the most suitable carbon source for growth and degradation of atrazine by strain ZF1.Secondly,sucrose and sodium citrate,and the degradation rate of atrazine in the treatment group with glucose as carbon source was the lowest,but their degradation rates all exceeded 90%.After 3.5 h of incubation,atrazine in all the treatment groups was completely degraded.The degradation rate of atrazine in the treatment group without adding carbon source was only lower than that in the starch treatment group,and the removal rate of atrazine reached 99.69%.
The results of the effects of additional nitrogen source on the growth and degradation of atrazine of strain ZF1 are shown in Fig.6e.After 2 h of incubation,the degradation rate of atrazine (100 mg · L-1)in all the treatment groups was over 90%.Among them,the concentration of strain ZF1 and the degradation rate of atrazine increased in the treatment group with urea,ammonium salt and nitrate;after 3.5 h of cultivation,atrazine was completely degraded in all the treatment groups,indicating that the addition of nitrogen source had a promoting effect on the growth of strain ZF1.
Discussion
ZF1,isolated from the experimental soil,which almost completely removed 100 mg · L-1atrazine after 2 h when the starch was used as carbon source,had greater atrazine-degrading ability than other atrazine-degrading microorganisms,such as strains SYSA (Zhanget al.,2008),YQJ-6 (Zhuet al.,2021) and ATR-Z5 (Zhuet al.,2020),which degraded 87% of the atrazine(20 mg · L-1) within 146 h,99.2% of the atrazine(50 mg · L-1) within 7 days,97.2% of the atrazine (100 mg · L-1) within 6 days,respectively.
To date,the known genes which encoded the enzymes to hydrolyze atrazine wereatzA,atzB,atzC,atzD,atzE,atzF,trzDandtrzN(Mulbryet al.,2002;Fabriceet al.,2006).Among them,atzAandtrzNencoded atrazine chlorohydrolase (Toppet al.,2000;Wackettet al.,2002).Next,atzBandatzCencoded hydroxyatrazine ethylamine-hydrolase and N-isopropylamide isopropylamine hydrolase respectively to hydrolyze hydroxyatrazine to cyanuric acid (Wanget al.,2013).Some bacteria degraded cyanuric acid to NH3and CO2,required the enzymes encoded byatzD/trzD,which acted in concert with those encoded byatzEandatzF(Toppet al.,2000;Meyeret al.,2009).In the present study,trzN,atzBandatzCwere amplified,and the genetic potential was close to that of strain TC1 (Sajjaphanet al.,2004).Based on the findings and previous reports,the following atrazine degradation pathway inPenarthrobacter ureafaciensZF1 could be proposed:atrazine hydro-xyatrazine-N-iso-propylammelidecyanuric acid.This pathway was similar to that in many other strains (Hanet al.,2009;Liet al.,2014).
The reported suitable temperature range ofAcinetobacter lwoffiiDNS32 was 25℃-30℃ (Guoet al.,2012),and that ofEnrerobactersp.was 25℃-35℃ (Liuet al.,2016).In this study,strain ZF1 had a wide temperature range (5℃-50℃),which still maintained a degradation rate of more than 80%under low temperature (10℃) or high temperature(50℃) within 3.5 h.Similarly,the growth and atrazine-degradation efficiency of strain ZF1 were acceptable at pH 5.0-11.0,demonstrating that pH range for atrazine biodegradation by ZF1 was wide and alkaliphilic.In fact,most previous strains could only degrade atrazine at low concentrations (Daiet al.,2007;Zhaoet al.,2018).Conversely,ZF1 had strong tolerance to atrazine and could tolerate atrazine at least at a concentration of 1 500 mg · L-1.In addition,the addition of nitrogen source had a promoting effect on the growth of strain ZF1.Some previous studies showed that additional nitrogen sources had a negative impact on atrazine degradation,such asPseudomonasADP (Desittiet al.,2016) and M91-3(Gebendinger and Radosevich,1999).In brief,strain ZF1 had strong tolerance to low temperature,high temperature and alkaline conditions,indicating that the strain had great potential for the remediation of atrazine-contaminated sites,particularly in cold,hot and alkaline environments.
Conclusions
In this experiment,ZF1,a bacterial strain,capable of highly degrading atrazine was isolated from agricultural soil,which was identified asPenarthrobacter ureafaciens.It was found that the strain ZF1 harboredtrzN,atzBandatzCgenes,which could transform atrazine to cyanuric acid.This strain ZF1 had a strong ability to biodegrade atrazine with a high efficiency of 50 mg · L-1· h-1.At the same time,strain ZF1 had strong tolerance to low temperature,high temperature and alkaline environment,indicating that strain ZF1 was suitable for bioremediation of atrazine contaminated sites in cold,hot and saline alkali areas.In conclusion,the isolation of strain ZF1 enriched the strain resources of atrazine degrading strains in middle-area of China,and it would have good application values in complex ecological environment.
 Journal of Northeast Agricultural University(English Edition)2022年4期
Journal of Northeast Agricultural University(English Edition)2022年4期
- Journal of Northeast Agricultural University(English Edition)的其它文章
- Journal of Northeast Agricultural University (English Edition)Instruction to Authors
- Total Contents of Volume 29(2022)
- Research Progress of Vitamins on Muscle Regeneration
- Real-time Prediction Model of Amount of Manure in Winter Pig Pen Based on Backpropagation Neural Network
- Investigation and Analysis of Grassland Plant Germplasm in Zhalantun City of Eastern Inner Mongolia
- Regulation of Migration,Phagocytosis and Apoptosis of Human Neutrophils by Recombinant Human Intestinal Alkaline Phosphatase
