Analyses of the differential expression of cloned Caspase-3 and Caspae-9 genes, of Onchidium reevesii, exposed to extreme hot and cold conditions
Fengshen Zhao, Heding Shen,*
aKey Laboratory of Aquatic Germplasm Resources Exploration and Utilization of Ministry of Education, Shanghai Ocean University, Shanghai, 201306, China
bNational Experimental Teaching Demonstration Center of Aquaculture Science, Shanghai Ocean University, Shanghai, 201306, China
cClassification and Evolution of Marine Animal System, Shanghai Ocean University Key Laboratory of Shanghai Universities, Shanghai, 201306, China
ABSTRACT
During low tide intertidal organisms face severe temperature fluctuations. This situation may be more pronounced in the context of climate change. To investigate the molecular response mechanism of Onchidium reevesii when stressed by hot and cold temperatures, we cloned its Caspase-3 and Caspase-9 genes and detected their expression characteristics using RT-qPCR. The results showed that the full-length of the cDNA of Caspase-3 consist of 1831 bp, and included an open reading frame (ORF) of 894 bp, a 5′UTR of 155 bp and a 3′UTR of 300 bp, and encoding for 297 amino acids. The full-length of the cDNA of Caspase-9 consisted of 2425 bp, and included an ORF of 1373 bp, a 5′UTR of 290 bp, and a 3′UTR of 759 bp, and encoding for 457 amino acids.During the evolution of species the two genes have been greatly conserved. A homology analysis and phylogenetic tree reconstruction revealed that O. reevesii is most closely related to Aplysia californica. The RT-qPCR showed that both genes were expressed in all of the tissues and that expression was highest in the hepatopancreas. It is speculated that these genes play an essential role in the immunity of O. reevesii to certain diseases.Conditions of temperature stress (40 °C and 10 °C) showed that both heat and cold stresses could induce significant changes in both genes.
Compared to the control group, under cold stress conditions the levels of expression of Caspase-3 and Caspase-9 were significantly upregulated (P <0.05) in each experimental group. When exposed to a constant temperature of 10 °C, their expression peaked after 1 h (4.32-fold, P < 0.05; and 5.58-fold, P < 0.05, respectively) and was at its lowest after 12 h (1.93-fold, P < 0.05; and 2.35-fold, P < 0.05, respectively). Their expression was upregulated after 24 h (2.68-fold, P < 0.05; and 4.21-fold, P < 0.05, respectively). The expression of both genes showed similar trends. Under heat stress, the expressions of the two genes showed a wave pattern. When exposed to a constant temperature of 40 °C, their expression was up-regulated after 1, 6, and 24 h, and down-regulated after 3 and 12 h. Except at 24 h, the other experimental groups did not differ significantly (P >0.05) from the control group. When subjected to various stress conditions the two genes displayed different expression patterns,indicating that the species has different strategies for coping with temperature changes. It further indicates that the effect of temperature on the species may play a role through the Caspase-dependent pathway. These may help to better understand the response, at the molecular level, of O. reevesii to conditions of hot and cold stress.
ARTICLEINFO
Keywords:
Onchidium reevesii
Caspase-3
Caspase-9
Temperature Stress
1.Introduction
The abnormal climate change caused by human activities could induce an increase in extreme hot or cold weather conditions, which may have a powerful impact on allothermic animals (Li et al., 1997). As a key environmental factor affecting the homeostasis of allotherms, the temperature may not affect only the spread of germs and the dissolution of toxic substances, but also the normal physiological functions (e.g.development, reproduction and immunity) of organisms (Cherkasov et al., 2006; Koenigstein et al., 2016). The intertidal zone is considered to be a suitable model for studying the impact of climate change on the distribution of biodiversity and on ecosystems (Helmuth et al., 2002;Southward et al., 1995). Previous studies have shown that the perfect heat shock response in intertidal organisms allows them to survive over a wide range of temperatures (20C). However, intertidal organisms,especially organisms in the high tide zone, are as sensitive to changes in temperature as are polar animals, and can serve as good indicators of climate change (Tomanek, 2010).
Previous studies have shown that high and low temperatures can induce an increase of reactive oxygen species (ROS) in an organism. This promotes the expression of antioxidant enzymes and heat shock protein genes, thereby offsetting the adverse effects of oxidative stress (Bagnyukova et al., 2007; Tomanek & Zuzow, 2010; Wang et al., 2019; Wu et al., 2015). However, excessive stress, beyond biological tolerance limits, can lead to oxidative damage and induce apoptosis. In multicellular organisms, apoptosis is a key process actively maintaining tissue homeostasis and eliminating potentially harmful cells (Jacobson et al.,1997). Apoptosis is triggered when cell surface death receptors (such as Fas) bind to their ligands (external pathway) or Bcl2-family pro-apoptotic proteins causing mitochondrial outer membrane permeability changes (intrinsic pathway) (Green & Llambi, 2015). Both pathways focus on the activation of the caspase family which is ultimately responsible for cell breakdown. They all have the following common properties: they are all aspartate-specific cysteine proteases; they all have a conservative pentapeptide active site ‘QACXG’ (X can be R, Q or D); and their precursors are all zymogens known as procaspases (Fan et al., 2005). According to specific protein domains and their functions,this family can be divided into promoters Caspases and effectors Caspases. Promoters Caspases contain the death domain DED (caspase-8 and -10) or the Caspase recruitment domain CARD (caspase-2, -9, -1,and -11), which mediate their dimerization or recruitment to larger complexes to promote their activation (Shalini et al., 2015). Among them,Caspase-9
is a key “promoter” of the mitochondrial pathway and can activate downstream Caspase.Caspase-3
is the main “executor” and is responsible for the hydrolytic cleavage of many key cellular proteins(Wang & Lenardo, 2000).The Onchidiidae are widely distributed in the intertidal zone, except in the polar regions.Onchidium reevesii
(Mollusca, Gastropoda, Pulmonata, Onchidiidae) lives mainly in the high-tidal zone of the southeast coast and the southern Yellow Sea of China (Liu & Huang, 2018). The optimum temperature range is relatively narrow (20–28C) (Huang et al., 2004). This species may be used as a suitable model of allotherms to evaluate the impact of cold and heat stress on these organisms (Shu et al., 2017). In the present study, we clonedCaspase-3
andCaspase-9
using the RACE method and analyzed the expression patterns of these two genes under conditions of cold (10C) and heat (40C) stress.Studies of the molecular level response ofO. reevesii
to temperature changes provides a good reference for research on the adaptive responses of this species.2.Materials and methods
2.1.Animals
O. reevesii
were collected from Yancheng City, Jiangsu Province in June 2019. Animals were acclimated in the tank for 2 weeks at 25 ± 1C and fed corn starch once per day. Other factors such as salinity and relative humidity were controlled in the optimum range, excluding the influence of these factors (Huang et al., 2004).2.2.Full-length cDNA cloning of the Caspase genes
Total RNA was extracted using Trizol reagent (TaKaRa, China) in accordance with the manufacturer’s protocol. The quality of the RNA was detected by using 1.2% Agarose gel electrophoresis, and the RNA concentration was determined using a NanoDrop 2000 instrument(Thermo Scientific, USA). The RNA was then reverse transcribed into cDNA by HiScript Q RT SuperMix for qPCR (+g DNA wiper) Kit, and stored at −20C.
Two incomplete cDNA sequences ofCaspase-3
andCaspase-9
were obtained from a cDNA library of the hepatopancreas ofO. reevesii
. Based on these two sequences, we designed multiple primers to amplify the middle sequence ofCaspase-3
andCaspase-9
(Table 1). Rapidamplification of cDNA ends (RACE) was then performed to obtain the 3and 5ends of the chains by SMARTER®RACE5/3Kit (TaKaRa,Japan). The purified PCR products were ligated with pGEM-T vector(Promega, USA) to construct the recombinant plasmid. The plasmid was introduced intoE. coli DH5α
on a LB selective medium containing ampicillin (100 mg/ml), IPTG (200 mg/mL) and X-gal (20 mg/ml). and cultivated overnight at 37C. The positive clones were sequenced by Sangon, BIO, Shanghai, China, after Blue-White Screening and PCR detection of the colony. Finally, the sequences obtained were assembled into complete cDNA.
Table 1Primers used in the present study.
2.3.Bioinformatic analysis of the Caspases
The open reading frame (ORF) and amino acid sequences were predicted using ORF Finder (https://www.ncbi.nlm.nih.gov/orffinder/) on NCBI. The physicochemical properties of proteins were analyzed by ProtParam (https://web.expasy.org/protparam/). Prosite (https://prosite.expasy.org/) was used to analyze the domains of the Caspase protein. Signal 4.1 (https://www.cbs.dtu.dk/services/SignalP-4.1/) was used to predict potential signal peptide cleavage sites. TMHMM(http://www.cbs.dtu.dk/services/TMHMM/) was used to analyze the transmembrane domains of the proteins. Protein secondary and tertiary structures were predicted by Phyre2 and Pymol software. Multiple amino acid sequence alignments were performed with DNAMAN software. The phylogenetic tree was constructed using the Neighbor-joining(NJ) method in MEGA 6 software, with 1000 bootstrap replications, the other parameters used were the default values.
2.4.Experiment to determine the differential expression of the genes in different tissues
To assess the differential expression of the genes in different tissues,nine individuals were kept in a tank at room temperature (25 ± 1C).Seven tissues including ganglion, hepatopancreas, bisexual glands,dorsal, abdominal, foot skin and trophi, were collected and immediately frozen in liquid Nand stored at −80C until used.
2.5.Temperature stress experiment
One-hundred and eight healthy animals were selected and randomly allocated to the cold (10C) or hot (40C) stress groups and exposed to these temperatures for 0, 1, 3, 6, 12 and 24 h, respectively. At each point in time, the hepatopancreas was harvested from nine individuals in each of the two groups and used for the analysis of mRNA expression.Experimental animals were transferred to an incubator (Bio Multi incubator; NK Systems, Osaka, Japan) for the different temperature treatments. All tissues were immediately frozen in liquid Nand stored at −80C until use.
2.6.Analysis of the mRNA expression of Caspase by RT-qPCR
Primers for RT-qPCR were designed using Primer Premier 5.0 software, with β-action selected as the internal reference (Table 1). A realtime reverse transcript polymerase chain reaction (RT-qPCR) was performed with the QuantStudio 6-Flex Real-time PCR System (Applied Biosystems, Life Technologies Corporation, USA) in accordance with the manufacturers protocol. The 20 μL PCR reaction system contained 10 μL of 2 × ChamQ Universal SYBR qPCR Master Mix (Vazyme, China), 0.4 μL of each primer, 2 μL of cDNA and 7.2 μL ddHO. The two-step q-PCR reaction was followed: 95C for 30 s, 40 cycles of 95C for 10 s, and 60C for 30 s.
The 2method was used to calculate the level of expression of the two genes. Statistical analyses were performed using One-Way ANOVA in SPSS 20 software. Differences were considered significant at P <0.05. The figures were drawn using OriginPro 8.
3.Results
3.1.Analyses of the sequences of Caspase-3
The complete sequence of Caspase-3 (GenBank accession no.MN612052) consisted of 1381 bp and included an ORF of 894 bp, a 5UTR of 155 bp and a 3UTR of 300 bp. The 3UTR contained a typical polyadenylation signal (AATAAA) and Poly(A) tail (Fig. 1).

Fig. 1.Sequences of Caspase-3 (top) and Caspase-9 (bottom) genes, from O. reevesii, and their encoded amino acid sequences. Dark grey indicates the CARD domain.Light grey indicates the CASc domain. The start and stop codons are boxed; and underlined shows P20 and P10. Histidine active sites are marked by the orange boxes.Cysteine active sites are labeled by the green boxes. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)

Fig. 1. (continued).
The predicted molecular weight (MW) was 33.22 kDa, the theoretical isoelectric point (pI) was 5.78, and the molecular formula was CHNOSThe predicted protein domain had the highly conserved CASc domain at the C-terminus (amino acids 48–296) and no DED or CARD domain at the N-terminus. It belongs to the effector Caspase (Logue & Martin, 2008). The large subunit (P20, R55-E180) and the small subunit (P10, V202-E296) were located in the CASc domain.Caspase-3 is usually present in the form of an inactive proenzyme. After activation, the large and small subunits in the CASc structural domain of this gene are dissociated and reassembled into active enzyme in the form of a tetramer, which induces the occurrence of cell apoptosis (Sylwia et al., 2017). The histidine active site (HSRSDCFVCVILTHG) and the cysteine active site (KPKLFFIQACRG) were both in the P20 large submit (Fig. 1). There was no signal peptide and transmembrane region in the Caspase-3 sequence. Prediction of the secondary and tertiary structures found that Caspase-3 contains 7 α-helices and 5 β-sheets(Fig. 2).
3.2.Analyses of the sequence of Caspase-9
The complete sequence of Caspase-9 (GenBank accession no.MN636785) consisted of 2425 bp, including an ORF of 1373 bp, a 5UTR of 290 bp and a 3UTR of 759 bp, with a typical polyadenylation signal sequence (AATAAA) and Poly(A) tail, and encoding for 457 amino acids.
The predicted molecular weight (MW) was 51.81KD; the theoretical isoelectric point (PI) was 6.15. Analysis of the protein domain revealed that the CARD domain (AA at positions 1–92) and the CASc domain (AA at positions 165–448) were located at the N-terminus and C-terminus,respectively. The CARD domain can regulate the downstream Caspase cascade reaction through the mutual binding response of proteins, and the self-activation or activation of other corresponding procaspases(Sattar et al., 2003). The large subunit (P20, E172-L301) and the small subunit (P10, I360–P448) were located in the CASc domain. The histidine active site (HSQADCFVMFIFSHG) and the cysteine active site(KPKLFFIQACQG) were both in the P20 large submit (Fig. 1). No signal peptide and transmembrane regions were observed in Caspase-9 sequences. Prediction of secondary and tertiary structures found that Caspase-9 contains 5 α-helices and 9 β-sheets (Fig. 2).

Fig. 2.Prediction of the tertiary structure of the Caspase-3 (top) and Caspase-9(bottom) genes.
3.3.Multiple sequence alignment and phylogenetic analysis of Caspase-3 and Caspase-9 genes
The homology analysis revealed that the Caspase-3 protein was very similar to that of Aplysia californica (93%), Biomphalaria glabrata (93%)and Pomacea canaliculata (90%). The protein sequences of the selected species all have a highly conserved pentapeptide active site QACRG(Fig. 3). The reconstructed phylogenetic tree showed that the vertebrate and invertebrate Caspase-3 were clustered separately in two distinct groups. The Caspase-3 from O. reevesii was first gathered with a Caspase-3–like protein from Aplysia californica, and then grouped with other invertebrates (Fig. 4).

Fig. 3. (continued).

Fig. 3.Multiple sequence alignment of O. reevesii Caspase-3 (top) and Caspase-9 (bottom) compared with other species.

Fig. 4.The constructed phylogenetic tree based on Caspase-3 (top) and Caspase-9 (bottom) amino acid sequences.
The homology analysis of Caspase-9 from O. reevesii revealed that the protein sequences were very similar to other species. The Caspase-9 protein shared 99% of its amino acid sequences with Biomphalaria glabrata and 98% with Aplysia californica. The protein sequences of the selected species all have a highly conserved pentapeptide active site QACQG (Fig. 3). In the phylogenetic tree, vertebrate and invertebrate Caspase-9 were clustered separately into two distinct groups. The Caspase-9 of O. reevesii was first grouped with a Caspase-9 protein from Aplysia californica, and then grouped with other invertebrates (Fig. 4).
3.4.The distribution of Caspase-3 and Caspase-9 mRNA expression in different tissues
The RT-qPCR showed that Caspase-3 and Caspas-9 genes were expressed in all tissues (Fig. 5). The expression of both genes was highest in the hepatopancreas, followed by the bisexual glands and least expressed in the dorsal and abdominal tissues, with distinct tissue specificity.
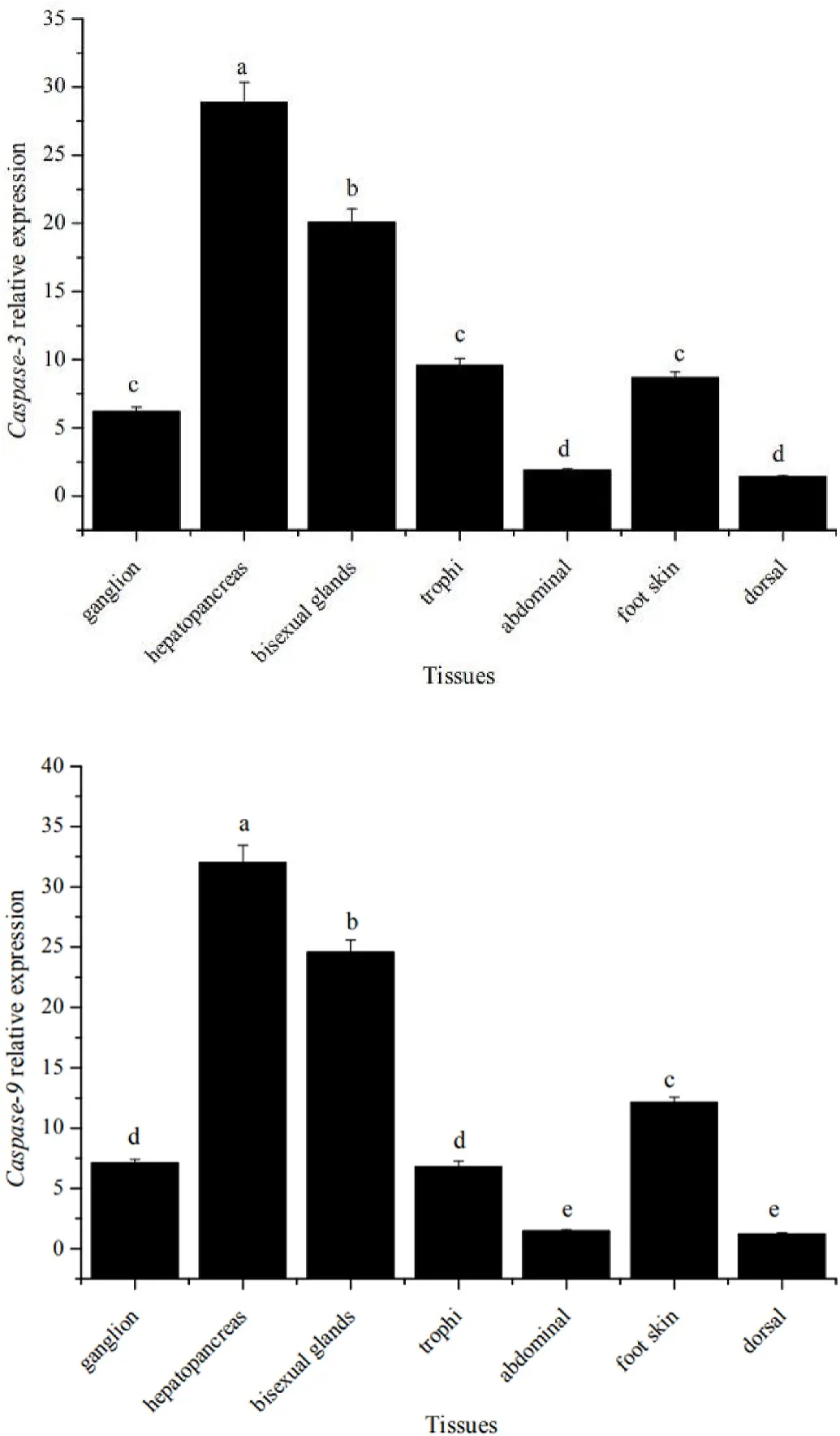
Fig. 5.Differential expression of Caspase-3 (top) and Caspase-9 (bottom) genes in the various tissues.
3.5.The effects of temperature on the expression of Caspase-3 and Caspase-9 mRNA
The expression of both genes at 0 h was used as the control. The expression of Caspase-3 and Caspase-9 genes were measured at 1, 3, 6, 12 and 24 h after exposure to high and low temperature conditions (Fig. 6).

Fig. 6.Expression of Caspase-3 (top) and Caspase-9 (bottom) under high and low temperature conditions. *shows the significant difference compared with the control group.
4.Discussion
Caspase is usually present in the cell as an inactive zymogen and is activated by specific proteolytic cleavage (Donepudi & Grütter, 2002).Its active form consists of a large subunit (17–20 kDa) and a small subunit (9–12 kDa). Each subunit contains residues essential for catalysis and substrate recognition (Stennicke & Salvesen, 2000). The catalytic domains of the Caspase family are very similar, but the N-terminus(prodomain) shows different characteristics in different types of Caspase genes (Ramirez & Salvesen, 2018).
The present study successfully cloned two apoptosis-related genes in O. reevesii. The CASc catalytic domains of both genes had P20 and P10 subunits. There were also two active sites (His and Cys) in the P20 subunit, and the pentapeptide sequences were in the Cys residues. The homology analysis and reconstructd phylogenetic tree revealed that the two genes were highly conserved during the evolutionary process. It showed that O. reevesii was most closely related to Aplysia californica.The difference between these two genes was that the pentapeptide sequence of Caspase-3 was QACRG, while for Caspase-9 it was QACQG.Caspase-9 also contained a CARD domain that Caspase-3 did not have.With the participation of cytochrome-c and dATP, this domain can bind to the N-terminal CARD of Apaf-1 to form a complex. Binding leads to the cleavage of Caspase-9, converting it to an active protease. Active Caspase-9 then activates downstream Caspase-3, thereby triggering apoptosis (Li et al., 1997).
Apoptosis is widespread in various tissues of the organism and is used to clear damaged or extra cells (Jacobson et al., 1997). In the present study, Caspase-3 and Caspase-9 genes were expressed in all tissues of O. reevesii, which is similar to that of Macrobrachium nipponense, Crassostrea gigas and Megahbrama amblycephala (Qu et al., 2014;Sun et al., 2017; Zhang et al., 2016). These indicated that the caspase apoptotic pathway is involved in multiple biological functions in both invertebrates and vertebrates. As the main immune organ of shellfish,the hepatopancreas plays an important role in the organism’s resistance to biotic and abiotic threats (Stennicke & Salvesen, 2000). Caspase-3 and Caspase-9 genes were highest expressed in the hepatopancreas of O. reevesii, suggesting that the two genes also play an indispensable role in the immune response of this species.
Studies on vertebrates and invertebrates have shown that abnormal environmental temperatures can trigger a series of physiological and biochemical reactions in organisms (Cooper, 2010; Tomanek & Somero,2000). Apoptosis also exists as a process in which the body adapts to environmental changes. Exposure to low temperature changed the physiological and immune response and induced oxidative stress and DNA damage of Takifugu obscurus. At the same time, apoptosis-related genes P53, Caspase-3, and Caspase-9 were significantly upregulated(Cheng et al., 2017). Heat stress also up-regulated the three apoptotic genes mentioned above, suggesting that the Caspase-dependent pathway may play an important role in cold and heat stress in Takifugu obscurus (Cheng et al., 2018). The expression pattern of apoptosis-related genes (Caspase-3, Caspase-9 and Bax), of Quasipaa spinose under cold stress, was faster than those under high-temperature stress; however, the specific reasons need to be studied further (Liu et al., 2018). Acute cold and heat stress caused DNA damage in Mytilus galloprovincialis and Mytilus trossulus, and induced time-dependent activation of Caspase-3 (Yao & Somero, 2012).
The caspase-dependent pathway refers to the process of apoptosis mediated by the Caspase family of genes (Zakeri et al., 2008). In the present study, we found that cold and heat stress can also induce changes in the expression of Caspase-9 and Caspase-3. This suggested that the Caspase-dependent pathway plays an important role in the apoptosis of shellfish, when induced by both cold and heat stress.Although both high and low temperatures play a role through the Caspase pathway, the expression profile of the same gene under different stresses was significantly different. We believe that the two genes are regulated differently under cold and heat stress, indicating that the organism has different strategies for coping with high and low temperatures. Compared with the control group, in the cold stressed groups the expression of mRNA of the Caspase-3 and Caspase-9 genes were significantly (P < 0.05) upregulated at each point in time. The most significant up-regulation was after 1 h (4.32-fold, P < 0.05; and 5.58-fold, P <0.05, respectively), then it gradually declined and was lowest after 12 h(1.93-fold, P < 0.05; and 2.35-fold, P < 0.05, respectively). After 24 h,the expression of both genes increased again (2.68-fold, P < 0.05; and 4.21-fold, P < 0.05, respectively). The trends in expression of the two genes were similar. Observations in the field found that with a decrease in temperature, O. reevesii began to look for “shelters” to help them avoid the cold weather. They are sensitive to low temperatures and poorly tolerant of it (Shen et al., 2011). Therefore, the Caspase-3 and Caspase-9 genes were immediately up-regulated after 1 h of exposure to cold conditions. After 24 h of exposure, they were up-regulated again,probably because the damaged cells produced by the low temperature needed to be removed to ensure the stability of the body’s environment.
Under high-temperature stress, the expressions of Caspase-3 and Caspase-9 genes were wave-shaped. After 1, 6, and 24 h of exposure to high temperatures they were up-regulated, and after 3 and 12 h of exposure they were down-regulated. No significant difference was observed between the other experimental groups and the control group(P >0.05) except for 24 h after exposure, indicating that this species is more resistant to high temperatures. This result is also in line with the living habits of O. reevesii when the weather turns warm. Previous studies showed that this species entered caves to avoid high and low temperatures (40C and 10C) (Shen et al., 2004, 2011, pp. 60–63).Therefore, we chose 10C and 40C as the lower and upper experimental temperatures.
O. reevesii becomes activity when the weather gets warm and “hibernate” when the weather becomes cold (Shen et al., 2004, 2011, pp.60–63). The experimental results, from the present study, confirmed that at a molecular level O. reevesii is more sensitive to low temperatures and less tolerant of them. In both the laboratory and in the field, the presence of intertidal crabs can help O. reevesii to survive better. One of the important reasons is that O. reevesii need the crab-excavated caves as“shelters” to successfully “overwinter” in (Shen et al., 2011). Studies have found that severe temperature fluctuations will reduce the breathing frequency of some intertidal organisms, such as crabs. It results in these species investing more energy in maintaining the steady-state of their bodies (Paganini et al., 2014), thereby reducing the energy used for other life activities such as growth and reproduction.Based on a long-term perspective, climate change will cause a decline in the population size of certain intertidal organisms, and even threaten the survival of certain species (Knight, 2014). The results of the present experiment also reflect that temperature changes, especially the evidence that low temperatures may harm the survival of O. reevesii.
5.Conclusion
The full-length cDNA of Caspase-3 and Caspase-9 in O. reevesii were obtained for the first time. The differential expression of mRNAs of Caspase-3 and Caspase-9 genes under hot and cold conditions suggested that temperature may induce apoptosis in O. reevesii through the Caspase-dependent pathway, however, the mechanism may be different.These results contribute to improving our understanding of the apoptotic mechanism of intertidal shellfish in response to temperature changes mediated by global climate change. It contributes information to formulating long-term sustainable strategies.
CRediT authorship contribution statement
Fengshen Zhao: Writing - original draft, Conceptualization, Investigation. Heding Shen: Resources, Conceptualization, Supervision.
Acknowledgments
This work was supported by the Construction Project of the Double First-class Disciplines of Fisheries.
 Aquaculture and Fisheries2021年6期
Aquaculture and Fisheries2021年6期
- Aquaculture and Fisheries的其它文章
- Overview of aquaculture systems in Egypt and Nigeria, prospects,potentials, and constraints☆
- microRNA expression profile offish erythrocytes
- Medaka gcnf is a component of chromatoid body during spermiogenesis
- Environmental and energy requirements for different production biomass of Nile tilapia (Oreochromis niloticus) in recirculating aquaculture systems(RAS) in Kenya
- Some reproductive and biometric features of the endangered Gangetic Leaf Fish, Nandus nandus (Hamilton, 1822): Implication to the baor fisheries management in Bangladesh
- The role of nitric oxide and neuronal nitric oxide synthase in zebrafish(Danio rerio) shoaling.
