Somatostatin 3 loss of function impairs the innate immune response to intestinal inflammation
Jing Ma, Jie Chen, Bruno Louro, Rute S.T. Martins, Aelino V.M. Canario,,*
aInternational Research Center for Marine Biosciences, Ministry of Science and Technology, Shanghai Ocean University, Shanghai, 201306, China
bKey Laboratory of Exploration and Utilization of Aquatic Genetic Resources, Ministry of Education, Shanghai Ocean University, Shanghai, 201306, China
cNational Demonstration Center for Experimental Fisheries Science Education, Shanghai Ocean University, Shanghai, 201306, China
dCCMAR-Centre of Marine Sciences, University of the Algarve, Gambelas Campus, 8005-139, Faro, Portugal
ABSTRACT
Somatostatin is a neuropeptide and a key regulator of the growth axis. Six Sst encoding genes (sst1 to sst6) have been identified in teleost fish genomes but little is known about their function. The present study aimed at replicating the context of the inflammatory bowel disease (IBD) and clarifying the involvement of sst3 in the intestine innate defence barrier in zebra fish larvae. We first established a CRISP/Cas9 sst3 deficient line (MT) and analysed the morphological and transcriptomic response to 0.4% dextran sulfate sodium (DSS). Alcian blue staining of larval sections 6 days post fertilization showed an increased in acidic mucins in the intestinal bulb and mid-intestine, with a much stronger response in the MT compared to wild type (WT). The transcriptomic analysis revealed that WT and MT shared enriched gene ontology (GO) terms and pathways linked to catabolism, chondrocyte development, innate immune system, xenobiotic metabolism and oxidative stress. In contrast, the WT specific response to DSS was enriched in GO terms and pathways linked to transcription and translation, various developmental processes, regulation of biosynthetic processes, apelin signalling and apoptosis while the MT specific response included terms and pathways linked to protein metabolism and catabolic processes, extracellular matrix – receptor interaction and proteasome and chondrocyte development. Overall, this study demonstrated that Sst3 deficiency impairs insulin growth factor and adipocytokine signalling exacerbating the inflammatory response to DSS.
ARTICLEINFO
Keywords:
Somatostatin 3
Zebrafish
IBD
Inflammation
RNA-Seq
1.Introduction
Somatostatin (SST) is a multifunctional hormone best known for its role in the hypothalamic control of growth hormone and pancreatic metabolic regulation (Brazeau et al., 1973; Hurley et al., 1994). SST localizes to the central nervous system (CNS), peripheral nervous system(PNS) and enteric nervous system (ENS), as well as in many organs and tissues (Patel et al., 1991). SST regulates the synthesis and secretion of many growth factors and hormones, including pancreatic polypeptide,ghrelin, leptin, insulin and glucagon (Boden et al., 1975; Dagogo-Jack,2015; Sakurai et al., 1974; Shimada et al., 2003). In addition to controlling the neuroendocrine system, there is evidence that SST receptors are expressed in innate and adaptive immune cells, macrophages, B and T lymphocytes, dendritic cells and monocytes (Aguila et al., 1991;Armani et al., 2007; Bhathena et al., 1981; Dalm et al., 2003; Ferone et al., 2012; Scicchitano et al., 1987). Furthermore, SST is able to inhibit the proliferation of T lymphocytes and granulocytes, and reduces the level of proinflammatory cytokines IFN-g (Casnici et al., 1997; Goetzl &Payan, 1984; Pawlikowski et al., 1985; Payan et al., 1984). Recently gene expression profiling of somatostatin knockout mice also suggested a possible role for somatostatin independent of growth hormone signalling in the innate immune response of the liver (Adams & Low, 2015).
SST is mostly stored in the digestive tract, especially in the gut and pancreas (Patel et al., 1981). SST is involved in the regulation of different processes along the digestive tract, including the main gastrointestinal functions, e.g. motility, secretion and absorption (Corleto, 2010). However, the intestinal epithelial cells are also the first layer of defence against food antigens, enteric bacteria and pathogens,releasing an array of pro-inflammatory chemokines and cytokines that trigger an inflammatory response (Jung et al., 1995). In order to control this process and avoid developing chronic inflammation and tissue damage, intestinal epithelial cells also secrete anti-inflammatory factors and neuropeptides, among them SST. SST stimulates secretion of cytokines (e.g. Th0, Th1 and Th2) and affects the secretion of pro-inflammatory cytokines from intestinal cells (Gonzalez-Rey et al.,2007; Levite, 1998). In addition to its effect on immunity cells and organs, somatostatin can also regulate the expression of various inflammatory factors. Somatostatin analogues can increase the expression of anti-inflammatory factor interleukin 10 (IL10) and inhibit that of pro-inflammatory factors, e.g. interferon gamma and tumor necrosis factor-alpha (TNF α) (Blum et al., 1992; Georgiadou et al., 2011).Conversely, inflammatory cytokines like transfroming growth factor β(TGF-β) can also inhibit SST release (Quintela et al., 1997).
The anti-inflammatory role of the somatostatinergic system has been associated to numerous human inflammatory diseases, mainly inflammatory bowel disease (IBD) (Ochsenkühn et al., 2003), and more recently in Sjögren syndrome, Graves’ disease and sarcoidosis (Signore,2015). IBD is a chronic condition of unknown etiology, in which patients display a chronic, relapsing intestinal inflammation resulting in intestinal lesions (Elson et al., 1994), leading to several pathological conditions, including ulcerative colitis (UC), Crohn’s disease (CD), and microscopic colitis (El-Salhy et al., 2017).
In IBD patients, the SST content in the intestine is significantly decreased (Koch et al., 1988), and the higher the grade of inflammation,the lower the number of SST-containing cells (Watanabe et al., 1992).Conversely, the number of SST receptor (SSTR) binding sites are significantly increased, namely in veins of inflamed intestines of IBD patients (Reubi et al., 1994).
The involvement of SST in these inflammatory states is primarily associated to the intestine innate immune response. In immunodeficient mice, sst1 controls innate immunity to Listeria monocyte genes (Boyartchuk et al., 2004). In macaques, SST is able to inhibit epithelial cytokine secretion (IL-8 and IL-1b) in normal physiological conditions and in Salmonella-triggered responses (Liu et al., 2015). Moreover, SST acts directly on tight junction proteins (Li et al., 2014), so that pre-treatment with SST can improve the barrier dysfunction of LPS-induced damage to tight junction (Lei et al., 2014). In addition, SST also exerts a protective barrier through its regulatory role on claudin-4 expression (Cai et al., 2018), suggesting that SST plays an important role in mucosal immune-homeostasis. For all these reasons, SST long-lasting analogue, octreotide (OCT), has been used to improve IBD symptoms in patients and animal models (e.g. murine). In this context,OCT has positive effects inflammatory bowel conditions, namely an anti-diarrheal effect (Yavuz et al., 2002), increases the intestine permeability (Cury et al., 2008), decreases mucosal damage through anti-inflammatory and anti-oxidant properties (Akgül et al., 2006; Wang et al., 2001). Nonetheless, how SST and synthetic analogues produce these effects and the precise mechanisms that are involved in its anti-inflammatory effect are far from understood.
The use of zebrafish Danio rerio as a model for intestinal damage is well established and the dextran sodium sulfate (DSS)-induced zebrafish IBD model mimics different features of mammalian IBD phenotypes,including neutrophilic inflammation in the intestine, excessive mucus production, increased intestinal lymphangiogenesis and upregulation of pro-inflammatory cytokines and markers ccl20, il1β, il23, il8, mmp9 and tnf-α (Oehlers et al., 2011, 2013, 2017; Okuda et al., 2015). SST is highly conserved in vertebrates and six Sst encoding genes (sst1-sst6) have been identified in zebrafish, (Liu et al., 2010). Although, a role for any of these genes in the inflammatory response of the fish intestine has not been demonstrated, there is evidence for a Sst-linked innate immune response. For example, in the intestine of turbot Scophthalmus maximus the number of Sst enteroendocrine positive cells increased in response to Enteromyxum scophthalmi (Losada et al., 2014); in rainbow trout Oncorhynchus mykiss skin, levels of Sst in mucus increased after exposure to parasite infection (Buchmann & Bresciani, 1997); and in the channel catfish Ichtalurus puntactus gills, sst2 transcription increased in response to infection with Flavobacterium columnare and decreased if fish were vaccinated (Zhang et al., 2017). In zebrafish, only sst1, sst3, sst4 are highly expressed in the intestine of adults (Sui et al., 2019) and during the first week post fertilization, when larvae rely solely on the innate immune system, sst1 is exclusively expressed in the central nervous system and pancreas while sst4 is expressed exclusively in the pancreas(Devos et al., 2002). This leaves sst3 as a likely candidate for gut expression in zebrafish larvae (Ng et al., 2005). Therefore, in the present study, we have used a CRISP/Cas9 sst3zebrafish line to evaluate the impact of loss of sst3 on intestinal inflammation of larvae triggered by DSS, as the most likely candidate to be potentially involved in this process.
2.Material and methods
2.1.Animal manipulations
The Shanghai Ocean University Experimentation Ethics Review Committee (under license SHOU-DW-2016-002) approved the methodology described in this study involving experiments with fish. Zebrafish were manipulated according to the procedures of the Institutional Animal Care and Use Committee of Shanghai Ocean University, Shanghai,China, to ensure the welfare and minimum suffering of the fish.
The AB zebrafish (Danio rerio) strain obtained from Shanghai Institute of Biochemistry and Cell Biology was used in this study. Zebrafish embryos were obtained from natural spawning and raised until 1 day post fertilization (dpf) in an incubator (Panasonic, MIR-254-PC) at 28.5C in recirculating water (60 μg/mL instant ocean sea salts) and supplemented with methylene blue up to 1 dpf. Larvae were grown at 28.5C for 5 days and from 6 to 15 dpf were fed with Paramecium spp.After 15 days, the fish were fed twice daily with newly hatched brine shrimp (Aquaneering-ZHBS-20-L Brine Shrimp Hatcher with Stand, San Diego, CA, USA) and maintained in a recirculating system (Aquaneering aquaculture system), with controlled water quality (temperature 26.3–28 0.5C, pH 7.24–7.89, Conductivity 500–700usμs/cm). The fish were maintained under a daily 10:14 (dark: light) regime with light intensity 54–324 lx.
2.2.CRISPR/Cas9 sst3 mutant
The ZiFiT Targeter software and the CRISPR/Cas9 gene editing system was used to generate zebrafish with mutations in exon 1 of the sst3 gene (Fig. 1A) (Sander et al., 2010). A set of specific primers (SST3-F and SST3-R, Table 1) containing the selected target site(GGCGTCTCGCGGCACTTCTG; Fig. 1B) were used to amplify the genomic region (GRCz11, chr15: 36115955–36120277). The specific gRNA was produced by polymerase chain reaction (PCR) amplification of pUC19-scaffold plasmid (Fig. 1B) by using the target site specific forward primer and the common reverse primer (respectively,T7-target-sfd and trac rev, Table 1). Each target sequence contains GG at the beginning and NGG as the PAM sequence. The gRNA was synthesized in vitro using the MAXIscript T7 Kit (Ambion) followed by digestion with DNase I (Ambion) for 15 min to remove DNA and purification with the mir Vana ™ miRNA Isolation Kit (Ambion). gRNA (100 pg) and Cas9 mRNA (300 pg) were co-injected (1 nl) into each single-celled embryo. Two days after injection, genomic DNA was extracted from three pools of 5 randomly taken embryos. Genomic DNA was PCR amplified using the primer pairs SST3-F and SST3-R (Table 1). Detection of mismatches was carried out with T7 endonuclease as recommended by the manufacturer (EnGen Mutation Detection Kit, New England Biolabs Beijing Ltd) and examined by agarose gel (2%) electrophoresis.PCR products were cloned into pMD19-T TA cloning vector (Takara),transformed into DH5α competent cells (Tiangen) and 20 independent colonies were selected for sequencing (Sangon Biotech, Shanghai,China).

Table 1Primer sequences used to produce sst3 mutants and in qPCR.
The founder fish carrying the Sst3 mutation were backcrossed with the WT to generate the heterozygous F(Fig. 1B). Fish from the Fgeneration carrying a 1 bp insertion and a 7 bp gene deletion were selected to produce the Fgeneration (Fig. 1C). All the Findividuals were raised together to avoid tank effects. The FWT and Sst3mutant fishes were crossed independently to produce the FWT and Sst3mutants used in the present study. Because of the DNA modification, Sst3mutants are only able to produce a 24 or 25 amino acids prepropeptide of Sst3, but not the active peptide as confirmed by sequencing (Fig. 1C).
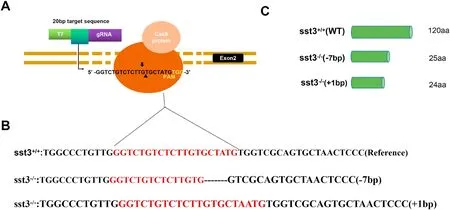
Fig. 1.CRISPR/Cas9 sst3 target and generation of F0 founders. A) structure of sst3 gene with target sequence indicated. B) DNA sequences of wild type and sst3−/−mutants; the sequences in red represent the inserted mutation. C) Schematic representation of the expected gene products from wild type and mutants. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
2.3.DSS treatment
Wild type (WT) sst3and mutant (MT) sst3were used to analyse the effect of DSS (Yeasen Biotech Co., Ltd.). Initial experiments were carried out to determine the DSS LD. WT embryos (n =26–32 larvae per concentration) were incubated for 72 h post fertilization (hpf)in sterile water before the tests. From a 10% DSS stock solution (w/v),dilutions were prepared (0.4%, 0.5%, 0.6%, 0.7%, 0.8%, 0.9% w/v) in triplicate in 24-well plates (1 mL per well) with the medium replaced daily. Each well had 8-10 larvae. After 96 h treatment, the mortality was scored and the LDcalculated using probit analysis (Bliss, 1934). The dosage with the least toxicity was used in subsequent experiments (0.4% DSS) also in 24-well plates (Supplementary file 1). In each experiment,the control (WT or MT in sterile water) and DSS (WT or MT treated with 0.4% DSS) groups comprised 6 wells containing 10 embryos/larvae per well and the experiment was repeated 3 times. Fertilized eggs from the same breeding pair were split between control and DSS groups and incubated for the first 72 hpf in sterile water. At 72 h the WT and MT DSS groups received the 0.4% DSS in sterile water for a further 72 h while the corresponding controls were incubated in parallel in sterile water only.At 144 hph (6 days) half of the larvae were used for histopathological analyses and the other half were used for RNA isolation.
2.4.RNA isolation and quantitative real-time PCR
Total RNA was isolated from 3 pools of 10 larvae per group using TRIzol (Invitrogen), according to the manufacturer’s instructions,quantified in the NanoDrop 2000C Spectrophotometer (Thermofisher)and run on agarose gel electrophoresis to test its integrity. For cDNA synthesis 1 μg total RNA and the PrimeScript™ RT reagent Kit were used together with gDNA Eraser (Takara) to ensure the removal of contaminant genomic DNA. The expression of sst1-sst6 (primers in Table 1) was detected by fluorogenic quantitative polymerase chain reaction (RT-qPCR) using the SYBRGreen I Master and the LightCycler 480 real time PCR detection system (Roche, Mannheim, Germany) according the manufacturer’s instructions. The β-actin gene, which showed no changes over the time course of the treatment, was chosen as reference gene to normalize the data. The qPCR efficiency was between 95% and 105%.
2.5.Histopathological examination
The larvae were fixed in Bouin’s solution (Sigma) overnight at 4C,which was followed by embedding in paraffin after dehydration by a series increasing alcohol grade solutions (70%–100%) and clearing with xylene. Sections (5 μm) were stained with Alcian Blue Periodic acid Schiff (AB-PAS Stain kit, Solarbio). Histological imaging was carried out on a ZEISS Imager M2 compound microscope with an Axiocam 506 color camera. The staining in the intestinal bulb and mid-intestinal area that contained mucus was quantified using the Image J software in each section.
2.6.RNA isolation, transcriptome assembly and pathway analysis
The total RNA from 3 pools of 10 larvae per group was isolated and run on agarose gels as described above. The purity of the RNA was assessed using the NanoPhotometer® spectrophotometer (IMPLEN, CA,USA) and the RNA concentration was measured using the Qubit® RNA Assay Kit in a Qubit® 2.0 Fluorometer (Life Technologies, CA, USA).RNA integrity was further assessed using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, USA). The RNA-seq libraries and sequencing were outsourced to Novogene.Sequencing libraries (3 μg total RNA/sample) were generated using NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (NEB, USA)following the manufacturer’s recommendations and index codes were added to attribute sequences to each sample.
Sequencing was carried on an Illumina HiSeq X Ten. Raw reads in fastq format were firstly processed through Novogene in-house perl scripts producing simple descriptive statistics and edited reads. Edited and cleaned reads were mapped to the zebrafish reference genome(zfish_GRCz11) with Galaxy (usegalaxy.eu) using the HISAT version 2.1.0 sequence aligner (Kim et al., 2015). The fasta and respective annotation file (.gtf) were downloaded from the Ensembl Genome Browser (http://ensembl.org). The estimated abundance of assembled and gene transcripts was generated with Stringtie version 1.3.6 (Pertea et al., 2016) and used to establish differential gene expression using DeSEQ2 version 2.11.40.6 (Love et al., 2014). The threshold for differential expression was P<0.05. Gene ontology (GO), KEGG and Reactome pathway enrichment analysis was performed using G. profiler v. e99_eg46_p14_f929183 (Raudvere et al., 2019) and enriched GO terms were summarized with ReViGO removing redundant GO terms(Supek et al., 2011) and displayed using ggplot2 in R. Sequences were deposited in ArrayExpress with accession E-MTAB-9018.
2.7.Statistical analyses
All data is expressed as mean ± standard deviation (mean ± SD).Two-way analysis of variance was used to compare sst1-6 expression during development and DSS treatments in WT and MT, Student’s t-test was used to compare the DSS LC50 between WT and MT. Statistical significance was accepted at P<0.05.
3.Results
3.1.Expression of sst1-6 in WT zebrafish between 48 and 168 hpf
RT-qPCR analysis of sst1-6 on WT whole larvae showed that all family members are expressed from 48 to 168 hpf (Fig. 2). Throughout this period, sst1-4 are the genes that display the highest expression with slight oscillations.

Fig. 2.Somatostatin genes relative expression to β-actin in zebrafish larvae of different ages. Similar letters indicate no statistically significant differences.
3.2.Effect of DSS treatment on survival
DSS at a dosage between 0.4 and 0.9% (w/v) caused differential mortality in zebrafish larvae. At concentrations above 0.5% the mortality rate was significantly increased in both WT (LD=0.676%) and MT (LD50 =0.591%) with significantly differences in LDbetween the two (two-tailed Student’s t-test, P =0.045, n =3) (Supplementary file 1).
3.3.General characterization phenotype sst3−/−mutant
The sst3mutant had no discernible anomalies and the size of adults for wild type and mutant at 140 days was statistically similar (P >0.05): respectively, length 22.9 ± 0.55 mm and 23.6 ± 0.50 and weight 2.2 ± 0.18 and 2.3 ± 0.14.
3.4.The effect of sst3 deficiency on intestinal inflammation
AB-PAS staining of untreated WT and MT larvae indicated no difference between the two groups on mucus production under normal physiological conditions (Fig. 3A–C). Once exposed to DSS, the intestine of WT larvae showed signs of mild inflammation as denoted by an increase of acid mucins staining and mucus secretion in the intestinal bulb and mid-intestine (P <0.05). However, in the MT there was a stronger response in acid mucin staining and mucus secretion (P <0.05) indicating that the absence of Sst3 aggravates the intestine inflammatory phenotype (Fig. 3A–C).
3.5.Transcriptomic response to treatment
The mRNAs from WT and MT control and treated fish were sequenced, and the quality of the raw data was assessed after filtering for contaminant sequences. The GC content of each library was similar with an average sequencing error rate of 0.03% (lower than 1%) with a Q20>90% and Q30 >80%. Altogether, we were able to map to the zebrafish genome 89.86%–91.29% of the clean reads. Most of the mapped genes in each sequencing library had low expression (43.32%–47.10%) and only 3.1%–3.5% of the genes were highly expressed (Supplementary file 2).
We found 874 differentially expressed genes (DEG) between WT controls and MT, 4262 DEG between WT control and WT treated with DSS (2521 of which exclusive of this comparison), 2746 DEG between MT control and MT treated with DSS (1112 exclusive), of which 1300 responded both to WT treated with DSS and MT treated with DSS (Fig. 4,supplementary file 3).
The top 10 up- and down-regulated DEG for each comparison is displayed, respectively, in Tables 2 and 3. Some of these DEG were simultaneously upregulated or downregulated in different treatments:ETS variant transcription factor 5 (etv5a_1) was upregulated in WT relative to MT and DSS-treated WT; zgc:136930 and type I cytokeratin,enveloping layer, like (cyt1l) were both upregulated in the WT and MT relative to the corresponding DSS-treated groups; sex hormone binding globulin (shbg_1) was downregulated in WT relative to both MT and DSS-treated WT; the lectin coding genes jacalin 9 (jac9) and jacalin 8 (jac8)were both downregulated in the WT and MT relative to DSS-treated WT and MT.

Table 2Top10 up-regulated differentially expressed genes with fold change relative to the second group in the comparison.
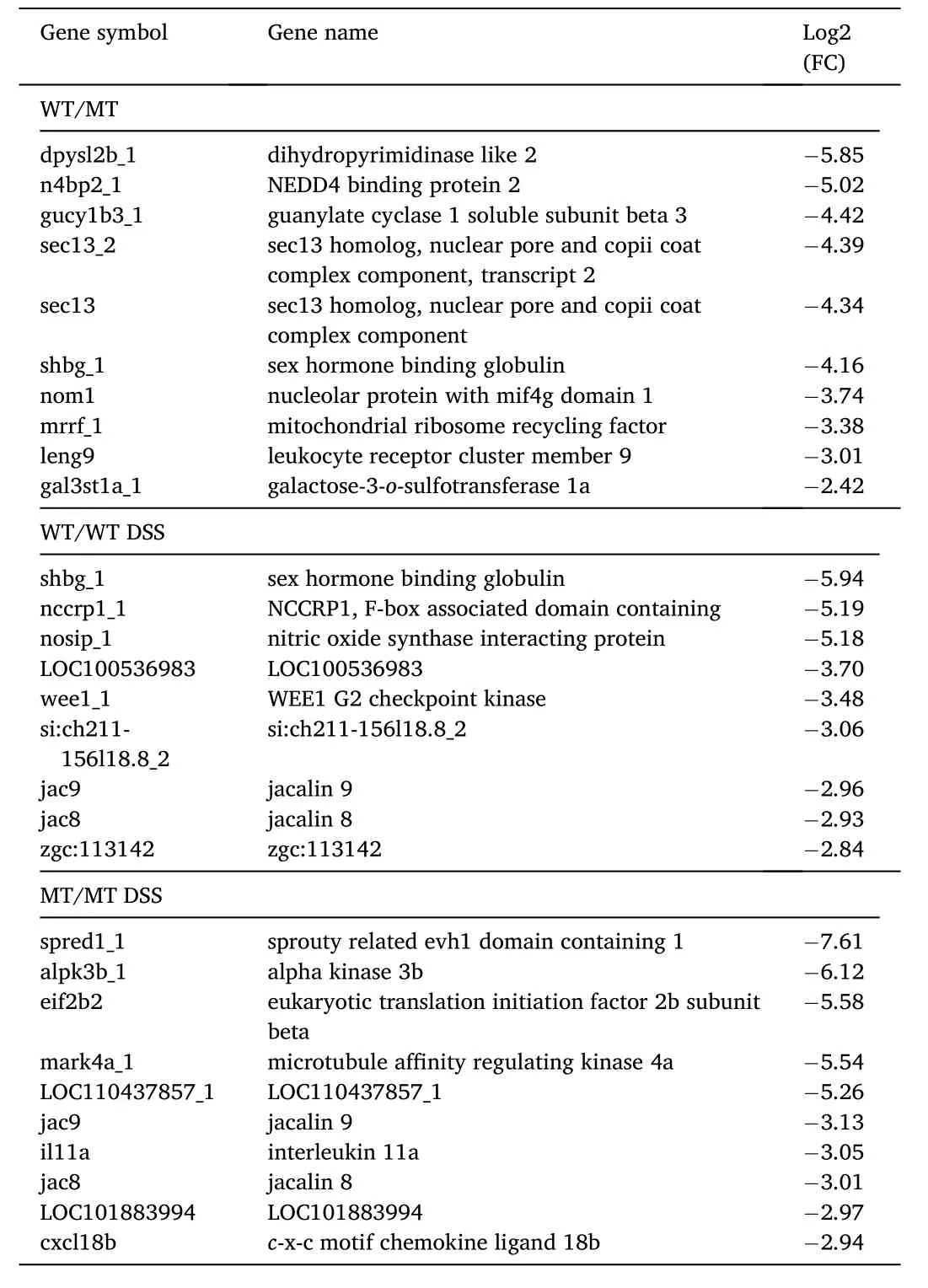
Table 3Top10 down-regulated differentially expressed genes with fold change relative to the second group in the comparison.
3.6.Gene ontology and pathways enrichment
The GO Molecular Function (MF) and Biological Process (BP) for DEGs resulting only from the sst3 knock out (control MT in relation to control WT) indicated, respectively 9 and 7 significantly enriched terms for oxidative metabolism, mitochondrial electron transport chain and ATP synthesis (Fig. 5A, Supplementary file 4). The KEGG pathway analysis for control MT in relation to control WT DEGs indicated only 2 significantly enriched pathways for oxidative phosphorylation and cardiac muscle contraction (Fig. 6A, Supplementary file 4). The Reactome analysis for the same DEGs only identified retinoid cycle in rods as significantly enriched.
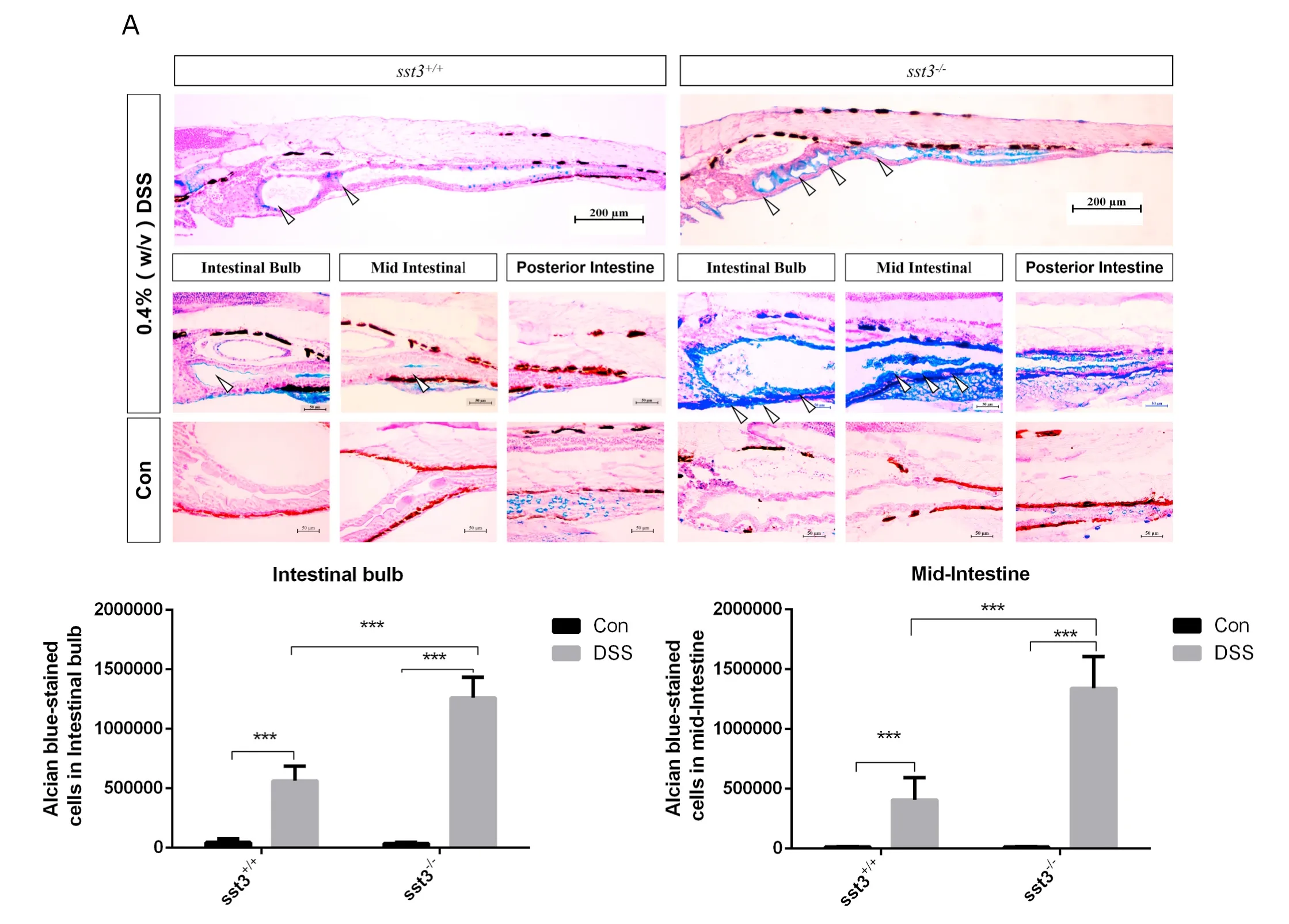
Fig. 3.- Effect of DSS administered between 3 and 6 dpf on the intestine of 6 dpf larvae. A) Alcian blue stained sections labelling acidic mucins in the digestive tract of DSS treated and untreated sst3+/+and sst3−/−larvae (upper panel). Amplified photographs of the intestinal bulb, mid and posterior intestine regions are shown in the lower panels. Open arrows point to stained regions in the anterior and mid-intestine. The intensity of Alcian blue staining was quantified in the intestinal bulb B)and mid-intestine C) was higher in DSS-treated than controls and in sst3−/− larvae compared to the sst3+/+. Asterisks indicate P <0.001. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
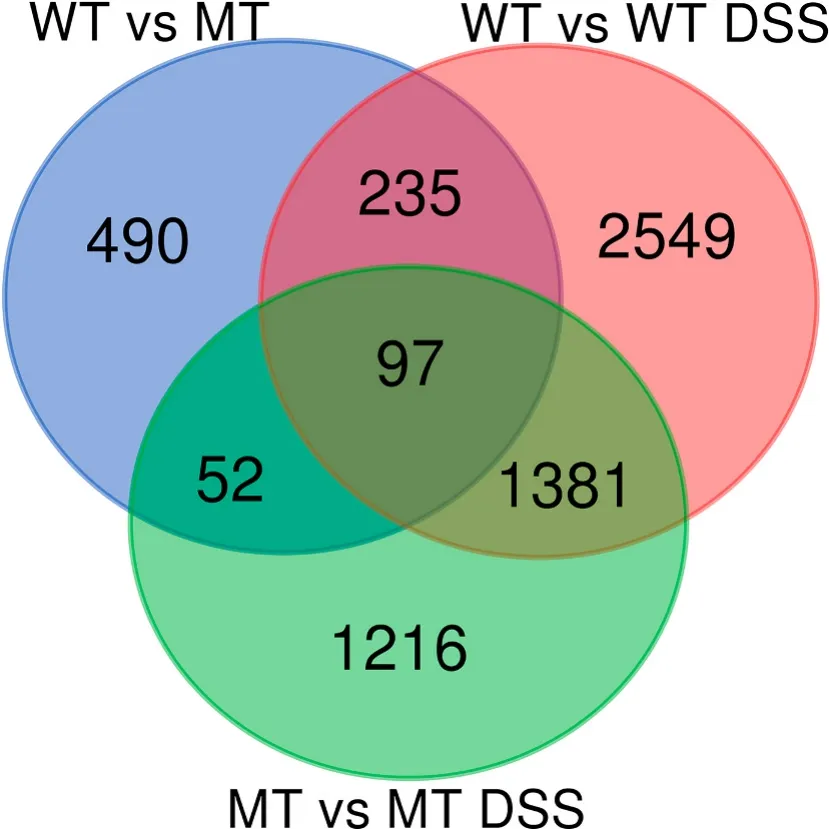
Fig. 4.Ven diagram indicating the number of differentially expressed genes exclusive or shared in the comparisons between WT and MT, without or with DSS treatment.
Exposure of WT to DSS significantly enriched 15 MF and 85 BP GO terms, mostly related to transcriptional regulation and activity, protein biosynthesis and catabolism, structural elements and different kinds of developmental processes (sensory organs, muscle, cytoskeleton)(Fig. 5B, Supplementary file 4). The same DEGs significantly enriched 9 KEGG pathways for ribosome, C-type lectin receptor signalling and apoptosis, xenobiotic metabolism, inflammatory and innate immune response (Fig. 6B, Supplementary file 4), while the Reactome was significantly enriched in 19 pathways for rRNA processing, mRNA translation and degradation, muscle contraction and protein degradation. The response of the MT to DSS significantly enriched 32 MF 24 BP GO terms several of which related to catabolic activity of proteins and lipids, glucose transport, antioxidant activity and detoxification(Fig. 5C, Supplementary file 4). The same DEGs significantly enriched 14 KEGG pathways mainly related to the proteasome, innate immune response, xenobiotic metabolism, oxidative stress and fatty acid metabolism (Fig. 6C). The Reactome enriched pathways were 51 for, among others, the innate immune system, oxidative metabolism, xenobiotic degradation and proteasome (Supplementary file 4).
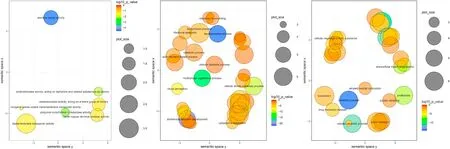
Fig. 5.–Scatterplots of Biological Function Gene Ontology terms after the redundancy reduction the in a two-dimensional space. Bubble color indicates P-value(legend in upper right-hand corner); size indicates the frequency of the GO term in the underlying GOA database (bubbles of more general terms are larger). A) DEGs between WT and MT, B) DEGs between WT and WT DSS, c) DEGs between MT and MT DSS. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)

Fig. 6.–KEGG pathways with significant enrichment. A) DEGs between WT and MT, B) DEGs between WT and WT DSS, c) DEGs between MT and MT DSS. The circle size is related to the proportion of genes represented from that pathway and the color represents the adjusted probability. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
When considering the shared response to DSS between WT and MT we find 19 significant MF and 9 BP GO terms, largely linked to catalyticactivity, glucose transport, catabolic processes, proteolysis and chondrocyte development. For the same DEGs we find 7 significantly enriched KEGG pathways related to the innate immune system, xenobiotic metabolism and oxidative stress (Supplementary file 4). When comparing the separate responses of the WT and MT to DSS, the WT specific DEGs included 8 MF and 88 specific BP GO terms linked to transcription and translation various developmental processes, regulation of biosynthetic processes and biosynthesis of macromolecules,while the MT-specific DEGS included 7 MF and 9 specific BP GO terms linked mostly to protein folding, protein metabolism and catabolic processes, extracellular matrix, and chondrocyte development (Supplementary file 4). The specific DEGs of the response of the WT to DSS were enriched in 4 KEGG pathways, e.g. ribosome, apelin signalling and apoptosis, and 21 specific Reactome pathways related to mRNA translation and degradation, while the DEG response of the MT to DSS enriched 3 specific KEGG pathways, e.g. extracellular matrix – receptor interaction and proteasome and 8 specific Reactome pathways related to collagen biosynthesis and trimerization, polyamine signalling and interleukin-1 signalling (Supplementary file 4).
3.7.Somatostatin signalling and growth regulation
Knock out of sst3 downregulated insulin-like growth factor binding protein 3 (igfbp3), retinol binding protein genes (rbp2a, rbp2b, rbp4) and upregulated insulin receptor a (insra) and insrb but did not affect other insulin signalling molecules. Treatment of WT with DSS downregulated npy, insulin (ins, ins_1), insulin-like growth factor binding protein, acid labile subunit (igfals), rbp2a, rbp2a_1, rbp4 and rbp4l, and upregulated growthhormone (gh1), leptin receptor (lepr_2) insrb, insulin-like growth factor a(igf1ra), igfr1b, like growth factor binding protein 1a and 1b (igfbp1a and igfbp1b). Treatment of MT with DSS downregulated igfbp2a, upregulated leptin b (lepb) and igfbp1a, and had no effect in other genes (Supplementary file 5).
4.Discussion
This study demonstrated that somatostatin 3 is important as an antiinflammatory factor in zebrafish and possibly other vertebrates. While knocking out sst3 causes significant changes in gene expression, mainly related to aerobic metabolism, induction of intestinal inflammation(enterocolitis) by DSS caused marked transcriptomic changes both in the WT and MT. However, the functional sst3 knock out was not able to mount the appropriate immune response to intestinal inflammation and these larvae died earlier than the WT controls. The transcriptome analysis unraveled the incapacity to regulate ins/igf signalling and adipocytokine signalling as possible causes of the exacerbated inflammatory response in MT.
The GO and KEGG enrichment analysis indicated that compared to the MT, the WT was highly enriched in processes related to oxidative metabolism (mitochondrial, electron transport, oxidative phosphorylation, and ATP synthesis and cardiac muscle contraction. Gh is the main regulator offish muscle growth via Igf (Fuentes et al., 2013) and our results are consistent with Sst regulation of the Gh-Igf axis in fishes(Bergan-Roller & Sheridan, 2018; Klein & Sheridan, 2008; Nelson &Sheridan, 2005). In fish, Sst stimulates lipolysis and carbohydrate mobilization (Eilertson & Sheridan, 1993; Kao et al., 1998) and inhibits oxidative metabolism and insulin secretion (Daunt et al., 2006) which is consistent with the upregulation of insulin receptor genes in the MT.Other members of the Gh-Igf axis were not affected by the mutation except for igfbp3 which was downregulated. Igf-binding globulins bind Igf, increase half-life and regulate binding to receptor in different tissues inhibiting mitogenesis, differentiation and survival, among other functions (Allard & Duan, 2018).
Of the 6 zebrafish sst genes, sst3 and sst4 are mostly expressed in the intestine of adult fish (Liu et al., 2010). As shown in the present study,both genes were already highly expressed from 48 to 168 hpf, as the digestive system of larvae develops. At 48 hpf the pancreas, liver,oesophagus and primordial intestine are bound as a functional unit(Wallace & Pack, 2003). At hatching (72 dpf) the intestine is already elongated onto a hollow tube, the mouth and anus opened to the exterior(Wallace & Pack, 2003) and the intestine is already colonized with microbes (Rawls et al., 2007). This suggests that the primary function of the intestine is as immune barrier prior to first feeding. In concert with the microbe colonization, at 96 dpf the gut mucosa already secretes enzymes (Pack et al., 1996; Wallace et al., 2005) and the enteroendocrine cells expressing somatostatin and glucagon (among others)are already present in the intestine. As during this period sst4 is exclusively expressed in the pancreas (Devos et al., 2002; Li et al., 2009), we hypothesized that sst3 could be involved in the regulation of the intestine immune barrier function prior to first feeding.
We used DSS to induce a phenotype of mild intestine inflammation on 6 dpf WT and MT zebrafish and found that the loss of function of Sst3 exacerbates the inflammatory response in the intestine, which is patent by the increase in acid mucins and neutrophilic infiltration in mutants.This phenotype mimics the chemically induced inflammatory phenotype of SSTR4 knockout mice, in which the lungs also presented increased inflammation, neutrophil/macrophage infiltration and mucusproducing goblet cells (Helyes et al., 2009). Thus, our sst3zebrafish and the SSTR4mice models demonstrate that somatostatin is a key anti-inflammatory neuropeptide in vertebrates.
As with the morphological response, the transcriptomic response of WT and MT fish differed. The DSS caused a larger transcriptomic response in the WT (4262 DEG) than in the MT (2746 DEG) suggesting limitations to mount the anti-inflammatory response in the MT.
Analysis of the GO terms, KEGG and Reactome pathways of the DEG in response to DSS shared by WT and mutants, showed that the innate immune system, xenobiotic metabolism and degradation were activated to cope with the inflammation. However, it is also clear that the loss of function of Sst3 conditioned the overall response to the inflammation by the MT and may have contributed to the lower mortality ED50 compared to WT. For example, in WT larvae, the apelin signalling is enriched. Apelin is an adipocytokine that regulates the intestine epithelial proliferation (Han et al., 2007) and goblet cell function (Wang et al., 2008). In inflamed intestines, apelin and its receptor are responsible for increasing the lymphatic drainage of the intestine and act in synergy with Il-10 to control the inflammation (Ge et al., 2018). The lack of activation of this signalling in mutants is in concert with the extreme inflammatory phenotype.
Conversely, the MT showed the enrichment of Il-1β signalling, which is consistent with the aggravated inflammatory phenotype, but also ECM-receptor and chondrocyte development, which was not enriched in the WT. The enrichment in ECM-receptor interactions may reflect the severity of the inflammatory phenotype as it is frequently accompanied by tissue damage and alteration of the structure of the intestine. In addition, recent studies have found that the balance of ECM synthesis and turnover is not only altered in IBD patients, it is also involved in mediating the inflammatory process. For example, hyaluronan overdeposition is associated with colitis (Kessler et al., 2015) and fibronectin and collagen increased deposition is associated to fibrosis in IBD(Kolachala et al., 2007; Lawrance et al., 2001). The enrichment in chondrocytes development in MT treated with DSS is also consistent with IBD. One of the phenotypes that concur with IBD in children is growth retardation. This condition is mainly due to the high Il-1β induced by inflammation that stimulates IGF1BP (Lang et al., 1996).Since IGF1 has higher binding affinity to IGF1BP than the IGF1R, the levels of active IGF1 (Lazarus et al., 1993) and acid labile subunit(Barreca et al., 1998) in the blood are reduced and impair GH signalling with the result that chondrocyte differentiation and bone development is delayed. The anti-inflammatory role, together with the potent stimulatory effect of SST on chondrocytes proliferation in mammals(Ferr´andez et al., 1992; Johansson & Madsen, 1987), suggest that in zebrafish sst3 innate immune function may also be essential for proper skeletal development.
The main counter mechanism to reduce Il-1 β signalling and inflammation is via activation of Il-10 signalling (Walter, 2014). However, IL-10 mutant mice, in which this signalling is inactivated, intestinal macrophages undergo abnormal differentiation and stimulate a hyperproduction of inflammatory cytokines, thus aggravating the inflammation (Shouval et al., 2014). The same pattern is also described in IBD patients with Chron’s disease (Kamada et al., 2008) and chronic colitis (Kamada et al., 2005). This proinflammatory tonus is also associated to splenomegaly in IBD patients (Kühl et al., 2007) and in DSS-treated mice models (Guri et al., 2011). In summary, although the inflammation was induced locally in the intestine, the lack of sst3 may also have resulted in a systemic immune response with life threatening consequences that replicates the most extreme complications of this inflammatory syndrome in IBD patients.
Interestingly, one of the top10 upregulated genes in the MT and in the DSS-treated WT relative to the WT was steroid hormone binding globulin (shbg). This binding protein is mainly produced in liver and is regulated by hormones and metabolic status. Low SHBG levels are a marker of metabolic syndrome and predictive of type 2 diabetes in mammals (Bonnet et al., 2009). However, there is still an ongoing debate on whether the low levels are due to the hyperinsulinemia or due to the effects of inflammatory cytokines implicated in these syndromes,e.g. TNFα and IL1β (Sim´o et al., 2012). In our study, ins and shbg had opposite responses in the DSS-treated WT, while in DSS-treated MT both ins and shbg did not respond indicating that the response of shbg and ins in inflammatory contexts is dependent on somatostatin signalling.
Immunity is also clearly linked to metabolism, and growing information shows that the adipose metabolism and secretion of cytokines(adipocytokines) are intimately related to inflammatory responses and autoimmune disorders. This is strongly supported by the fact that multiple inflammatory disease phenotypes are also associated with insulin resistance and/or obesity disorders (Guo, 2014). In our study, adipocytokine signalling is significantly affected in DSS-treated WT larvae but not in DSS-treated MT. For example, in mammals, leptin is an essential adipocytokine involved in the regulation of energy reserves and expenditure, inflammatory responses and in the brain central regulation of glucose homeostasis in concert with neuropeptide Y and agouti-related peptide (Mayer & Belsham, 2009; Palou et al., 2009). In fish, however, leprdid not show altered adipostasis but showed hyperinsulinemia and insulin resistance (Michel et al., 2016). Likewise,Npy Y1receptor deficient mice (Burcelin et al., 2001) also displayed hyperinsulinemia and insulin resistance which could be compensated by leptin administration. Another adipocytokine, the retinol binding protein 4 (RBP4), is also significantly increased in insulin resistance, type 2 diabetes mellitus and similar metabolic abnormalities (Yang et al.,2005).
Altogether, the work presented in this manuscript shows that the innate immune system of 6 dpf zebrafish larvae depends strongly on somatostatin signalling to cope with inflammatory stressors and demonstrates that Sst3 is an anti-inflammatory neuropeptide. WT zebrafish larvae respond to mild intestinal inflammation with a significant mucus secretion and apelin signalling possibly to control lymphatic drainage and inflammation. This is absent in the mutant in which Il-1β signalling,ECM-receptor and chondrocyte pathways are enriched indicating severe inflammation and possibly tissue damage. Sst3 loss of function revealed the incapacity of the innate immune system to cope with the DSS induced aggression to the intestine, ultimately resulting in an exacerbated inflammation and death of the mutants.
CRediT authorship contribution statement
Jing Ma: Conceptualization, Investigation, Writing - original draft.Jie Chen: Methodology. Bruno Louro: Investigation. Rute S.T. Martins: Writing - review & editing. Adelino V.M. Canario: Conceptualization, Resources, Supervision, Writing - review & editing.
Declaration of competing interest
The authors declare no competing interests.
Acknowledgements
This research was supported by the Shanghai Ocean University First-Class Disciplines Project of Fisheries, the open project of the International Research Center for Marine Biosciences (Ministry of Science and Technology) and the International Research Group Program at Shanghai Ocean University, and Portuguese national funds from FCT - Foundation for Science and Technology through project UIDB/04326/2020. RSTM was funded by FCT-Foundationfor Science and Technology- I.P., under the contracts Norma transit´oria- DL57/2016/CP1361/CT0021 with the University of Algarve.
Appendix A.Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.aaf.2020.09.001.
 Aquaculture and Fisheries2021年6期
Aquaculture and Fisheries2021年6期
- Aquaculture and Fisheries的其它文章
- Overview of aquaculture systems in Egypt and Nigeria, prospects,potentials, and constraints☆
- microRNA expression profile offish erythrocytes
- Medaka gcnf is a component of chromatoid body during spermiogenesis
- Environmental and energy requirements for different production biomass of Nile tilapia (Oreochromis niloticus) in recirculating aquaculture systems(RAS) in Kenya
- Some reproductive and biometric features of the endangered Gangetic Leaf Fish, Nandus nandus (Hamilton, 1822): Implication to the baor fisheries management in Bangladesh
- The role of nitric oxide and neuronal nitric oxide synthase in zebrafish(Danio rerio) shoaling.
