Mesenchymal stem cell-derived exosomes inhibit the VEGF-A expression in human retinal vascular endothelial cells induced by high glucose
Guang-Hui He, Ying-Xue Ma, Meng Dong, Song Chen, Yu-Chuan Wang, Xiang Gao,Bin Wu, Jian Wang, Jun-Hua Wang
1Clinical College of Ophthalmology, Tianjin Medical University, Tianjin 300070, China
2Tianjin Eye Hospital, Tianjin Eye Institute, Tianjin Key Lab of Ophthalmology and Visual Science, Tianjin 300020, China
3Ophthalmic Center of Xinjiang Production and Construction Corps Hospital, Urumqi 830002, Xinjiang Uygur Autonomous Region, China
4Medical College of NanKai University, Tianjin 300000,China
Abstract
● KEYWORDS: mesenchymal stem cells; exosomes;retinal vascular endothelial cells; vascular endothelial growth factor A; coculture
INTRODUCTION
Retinal neovascular disease has become a major cause of blindness due to eye diseases[1]. The vascular endothelial growth factor (VEGF), fibroblast growth factor(FGF) and interleukin-8 (IL-8) were shown to stimulate vascular proliferation[2]. VEGF is the most important cytokine that promotes retinal neovascularization. Anti-VEGF drugs targeting VEGF are widely used in clinical practice, but there are many limitations[3-4].
Mesenchymal stem cells (MSCs) are a type of pluripotent stem cell that shows multidirectional differentiation after continuous subculture and cryopreservation[5]. The most promising pluripotent stem cells for clinical applications are human umbilical cord mesenchymal stem cells (hUCMSCs),which have been used to treat diabetes[6]and bone formation[7],and cartilage injury-related diseases. However, due to the shortcomings of poor differentiation or tumorigenicity[8],hUCMSCs was restricted from being widely used in clinics.
Exosomes are natural information carriers among cells.Exosomes can transport proteins and RNA to target cells through shuttling among the cells to induce the target cells to respond accordingly. In previous research, we found that MSCderived exosomes can downregulate VEGF-A expression in retinal pigment epithelial (RPE) cells[9]. This research combined the two major research hotspots of human MSCs and exosomes, based on the existing researches, introduced MSCs exosomes into the research of anti-VEGF treatment of retinal neovascular diseases creatively, as to improve the treatment of stem cells to the nano level, reduce the side effects and risks of treatment. This study established a high-glucosegenerated model of human retinal vascular endothelial cells(HRECs) to observe the effects of hUCMSC-derived exosomes on the VEGF-A protein and mRNA levels of HRECs induced by high glucose and to investigate whether hUCMSC-derived exosomes have anti-VEGF effects.
MATERIALS AND METHODS
Ethical Approval The study was approved by the Tianjin Medical University Medical Ethics Committee and conducted in accordance with the Declaration of Helsinki, including current revisions, and with Good Clinical Practice guidelines.
Isolation and Identification of hUCMSC-Derived Exosomes hUCMSCs were cultivated in MSC medium (containing penicillin and streptomycin at 100 U/mL each; Gibco BRL,USA) containing 10% foetal bovine serum (FBS) in a 37°C and 5% CO2incubator, and changed the medium every 3 to 4d.When the hUCMSCs reached 85% confluence, the supernatant was discarded and 2 mL of 0.25% trypsin (Cat#:27250-018,Gibco BRL, USA) was added to the culture flask for cell digestion and then subcultured the cells. Well-growing cells were selected at generations 5 to 7 and cultured for 48h in a culture medium containing 10% Exo-FBS (exo-FBS, SBI,USA). Supernatants of the hUCMSCs were centrifuged at 300×, 2000×, and 10 000×g sequentially and filtered (0.45 µm).Then, a low-temperature ultracentrifuge (Beckman, USA) was used to centrifugete sample at 100 000×g at 4°C for 70min,we collected the precipitate and diluted it with phosphate buffered saline (PBS), and the sample was centrifuged again at 100 000×g for 70min and washed once. Finally,were suspended the collected exosomal concentrate in PBS containing 10% DMSO. The sample was sterilized with a 0.22 µm filter membrane and stored at -80°C.
The exosome sample was diluted at a ratio of 1:10, a copper mesh was dipped into a small amount of sample, and then, the copper mesh was placed in 3% phosphotungstic acid solution for 5min to deepen the background (negative staining). The sample was observed on a transmission electron microscope at 80 kV.
Western Blot Western blotting was used to detect the expression of hUCMSC-derived exosome-specific marker proteins. The exosome sample was mixed with 5× loading buffer (containing β-mercaptoethanol), boiled for denaturation for 5min, and placed in an ice bath for 5min. An appropriate amount of protein sample was loaded on SDS-denatured 10%polyacrylamide gel electrophoresis (SDS-PAGE) until the target protein was effectively separated. The membrane was placed into a Blotto containing the corresponding primary antibody overnight at 4°C. The membrane was placed in a Blotto containing the corresponding secondary antibody (HRPlabelled goat anti-rabbit antibody; anti-CD63, anti-CD9, anti-HSP70, anti-GRP94; Saier Bio, China), shaken and rinsed 4 times in total. After development with ECL developer solution and exposure of the photosensitive band, the development and fixation processes were carried out. The band was imaged and analysed by LabWorksTM (UVP, USA). GRP94 was used for comparison.
Nanoparticle Tracking Analysis Exosomal samples were diluted with 1×PBS buffer. A ZetaView PMX 110 nanoparticle tracking analyzer (NTA; Particle Metrix, Germany) and the corresponding ZetaView 8.04.02 software was used to analyse the diluted sample to measure the particle size and concentration of the sample. NTA measurement values at 11 locations were recorded and analysed, and 110 nm diameter polystyrene particles were used to calibrate the Zeta View system and maintain the temperature at 23°C-27°C.
Cell Grouping and High Glucose-treated HRECs HRECs(ATCC, USA) were routinely cultured in EGM2-MV medium(Lonza, USA) containing 10% FBS in a 37°C cell incubator.The culture medium containing 30 mmol/L glucose EGM2-MV was replaced in the high glucose group in a hypoxic cell incubator containing 1% oxygen. Pictures were taken under an inverted microscope atr 0h, 24h, and 48h of incubation. In this study,HRECs cultured under conventional conditions were defined as the control group (group A). HRECs in the Transwell coculture system, in which the upper layer had HRECs with high glucose and the lower layer had a certain volume of PBS added to the culture medium, were defined as group B.The upper layer of the Transwell coculture system of group C also had high glucose-treated HRECs, and the lower layer had culture medium supplemented with hUCMSC-derived exosomes, with a concentration of 25 µg/mL. The conditions of group D were the same as those of group C, and the concentration of hUCMSC-derived exosomes was adjusted to 50 µg/mL. The concentration of hUCMSC-derived exosomes in group E was titrated to 100 µg/mL.
Flow Cytometric Analysis of Proliferation Flow cytometry was carried out after 24h of cocultivation. The reaction mixture was prepared according to the instructions of the EdU-488 cell proliferation detection kit (Biyuntian Bio, China), which contained 2.5 mL of click reaction buffer, 2.15 mL CuSO4,100 µL CuSO418, 5 μL Azide 488, and 250 µL click additive solution. Then, 0.5 mL of reaction solution was added to the culture plate and detected on a flow cytometer.
Cellular Immunofluorescence After 24h of cocultivation,immunofluorescence were performed. The 300 μL of 4%paraformaldehyde was added at room temperature for 30min.After the cells were washed with PBS three times, 300 μL of 0.05% Triton-X-100 (diluted in PBS) was added for permeabilization. Then, 300 μL of 10% donkey serum was added at room temperature and 300 μL of diluted anti-VEGF-A primary antibody binding solution in 1% donkey serum was added overnight at 4°C. Then, 300 μL of 1% donkey serumdiluted secondary antibody (Saierbio, Tianjin) was added to each well. After incubation and washing, 300 μL of DAPI(1:1000 dilution, final concentration 1 μg/mL) was added, 3 μL of fluorescence protection agent was added dropwise, and the fluorescence was observed under a fluorescence microscope.
Real-time Quantitative Polymerase Chain Reaction Experiment According to the instructions of the Trizol RNA extraction kit (Invitrogen, USA), the total RNA of the cells was extracted and stored at -80°C. Premier 5.0 software was used to design upstream and downstream primers. VEGF-A:upstream primer 5’-GGCTGTTCTCGCTTCG-3’, downstream primer 5’-TGTCCACCAGGGTCTCG3-3’; the amplified fragment length was 544 base pairs (bp). β-tublin: upstream primer 5’-ATCCCATCACCATCTTCC-3’, downstream primer 5’-ATCACGCCACAGTTTCC-3’; the amplified fragment length was 380 bp. The RT reaction system is as follows: 3 µL total RNA, 1.0 μL OligodT primer (100 pmol/μL), and up to 13.5 μL with DEPC water. The reaction was mixed well and centrifuged for 5s. The sample was denatured at 65°C for 10min and incubated in an ice bath for 2min. The following reagents were added: 4μL 5×buffer, 1 µL dNTPs (10 mmol),0.5 µL RNasin (40 U/μL), and 1 µL M-MLV (200 U/μL). The M-MLV reverse transcriptase, RNase inhibition reagent, and SYBR Premix Ex Taq kit reagents (TaKaRa Company, Japan)were mixed, immediately centrifuged, incubated at 42°C for 1h , 70℃ for 10min. The quantitative PCR system was as follows: 2×SYBR Premix Ex Taq 10.0 μL, PCR sense primer(5 pmol/μL) 1.0 μL, PCR antisense primer (5 pmol/μL) 1.0 μL,template (RT product cDNA) 1.0 μL, and DDW supplement to 20 µL; the thermocycler reaction was as follows: 94°C for 30s,58°C for 30s, and 72°C for 30s for 40 cycles. An iQ5 Real-Time PCR Amplifier (Applied Biosystems, USA) was used for detection. The data analysis method was as follows: fold change=2-ΔΔCt, ΔΔCt=(Ct1-Ct2) - (Ct3-Ct4); Ct1 is the critical cycle number of the gene to be tested (VEGF-A) in the sample from the treatment group. Ct2 is the critical cycle number of the housekeeping gene (β-actin) in the sample from the treatment group. Ct3 is the critical cycle number of the gene to be tested (VEGF-A) in the control sample. Ct4 is the critical cycle number of the housekeeping gene (β-actin) in the control sample.
Statistical Analysis The results of cell immunofluorescence were quantitatively analysed using Image-Pro Plus software.The data obeys the normal distribution and accepts test for homogeneity of variance. It was presented as the mean±SEM.All experiments were repeated at least three times. SPSS 13.0 statistical software (SPSS, Inc., Chicago, USA) was used for comparisons among multiple groups by one-way ANOVA, and at-test was used for comparisons between two groups.P<0.05 was considered significant.
RESULTS
Characterization of MSC-derived Exosomes Under transmission electron microscopy, the diameters of exosomes derived from hUCMSCs were diverse. The diameters were approximately 50-200 nm. The membrane structure was visible on the periphery of the vesicle. The interior of the cavity showed low electron density components, and the background was clear without contamination (Figure 1A).
The Western blot results confirmed the expression of the markers CD9, CD63, and HSP70 on the surface of vesicles,and the expression of the marker GRP94, which was used as an indicator of contaminants, was negative (Figure 1B).
The isolated exosomes were diluted 6400 times, and then, the particle size of the obtained exosomes was determined by NTA technology. The results showed that the average particle size of the hUCMSC-derived exosomes was 122.7 nm (Figure 2).
High Glucose-cultured HRECs The cells had a paving stone-like morphology at the beginning, with the extension of the culture time, the cells under high glucose gradually changed from a paving stone appearance to a spindle shape, the cell polarity disappeared, and the gap between cells increased(Figure 3).
Flow Cytometry The proliferation rate of the HRECs induced by high glucose increased slightly compared with that of the HRECs cultured in normal glucose. After coculture with the hUCMSC-derived exosomes for 24h, the proliferation rate of the HRECs decreased with increasing exosome concentration and gradually approached that of the blank control group (there was no significant difference in the proliferation rate of each group,P>0.05), suggesting that cocultivation with exosomes had no significant effect on the proliferation rate of HRECs(Figure 4).
hUCMSC-derived Exosomes Downregulated the VEGF-A Level In the Glucose-induced HRECs Immunofluorescence results demonstrated that after the high glucose-induced HRECs were cocultured with the hUCMSC-derived exosomes for 24h, the intracellular VEGF-A level was downregulated,and the magnitude of the downregulation was proportional to the cocultured exosome concentration within a certain range(Figure 5). The relative level in each group was significantly different (F=39.03,P<0.01).
The level of VEGF-A mRNA in the high glucose group was significantly higher than that in the control group (t=6.928,P<0.01). Consequently, the mRNA levels of VEGF-A were markedly decreased in the hUCMSC-derived exosome-treated HRECs at 8, 16, and 24h compared to those of the model group with the duration of treatment time (8h:F=7.253; 16h:F=16.98; 24h:F=22.62,P<0.01). In the same time, the higher the concentration of hUCMSC exosomes added, the greater downregulation the VEGF-A mRNA expression in the high glucose-treated HRECs was (group C:F=4.137; group D:F=13.64; group E:F=22.19,P<0.05; Figure 6).
The level of VEGF-A protein in the high glucose induction group was significantly higher than that in the control group(t=15.57,P<0.001). The protein levels of VEGF-A in HRECs were evaluated in response to the hUCMSC-derived exosome treatment and were found to be markedly decreased in the hUCMSC-derived exosome-treated HRECs at 8, 16, and 24h compared to those of the model group with the duration of treatment time (8h:F=9.45; 16h:F=12.86; 24h:F=42.28,P<0.05). In the same time, the higher the concentration of the hUCMSC-derived exosomes added, the greater downregulation VEGF-A expression in the high glucose-treated HRECs was(group C:F=7.96; group D:F=17.29; group E:F=11.89,P<0.05; Figure 7).
DISCUSSION
Exosomes are biological nanovesicles with a diameter of approximately 30-1100 nm secreted by cells. They exist in almost all mammalian cells and are responsible for communication between cells[10]. These vesicles are closely related to the immune response, cell proliferation,inflammation, lactation and neuronal function[11-15]and are involved in different stages of progression of various diseases,such as liver disease[16], neurodegenerative disease[17]and severe cancer-related diseases[16]. The composition of exosomes is complicated. Currently, 4563 kinds of proteins, 764 kinds of miRNAs, 1639 kinds of mRNAs and 194 kinds of lipids have been detected in exosomes.
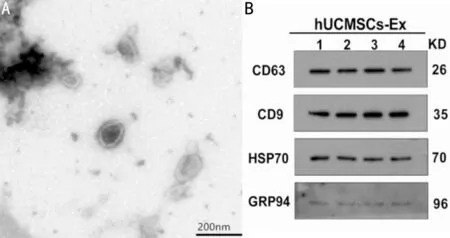
Figure 1 The membrane structure of the hUCMSC-derived exosomes under transmission electron microscope.
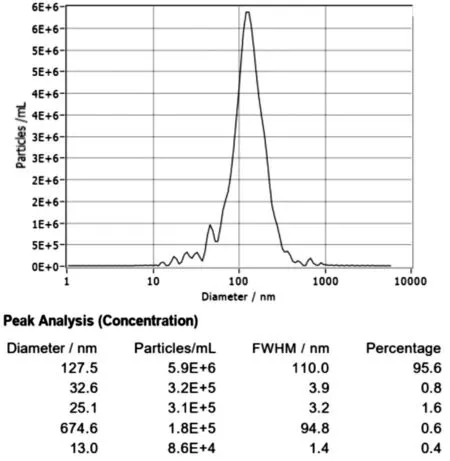
Figure 2 Nanoparticle tracking analysis of the hUCMSC-derived exosomes.
Current techniques for separating exosomes include ultracentrifugation, ultrafiltration or size exclusion chromatography (SEC), polymer precipitation, immunoaffinity capture technology, and the recently emerging microfluidic technology. Among them, ultracentrifugation technology is the most commonly used and considered to be the “gold standard” method for separating exosomes[18]. Transmission electron microscopy is the most commonly used technology to characterize exosomes[19]because it provides information on the morphology and structure of exosomes, composition information,etc.Moreover, this technique provides direct observations of the structure and morphology of exosomes of different sizes[20-21].

Figure 3 High glucose-cultured HRECs.

Figure 4 Flow cytometry to detect the cell proliferation rate of HRECs in each group when cocultured with the hUCMSC-derived exosomes for 24h.
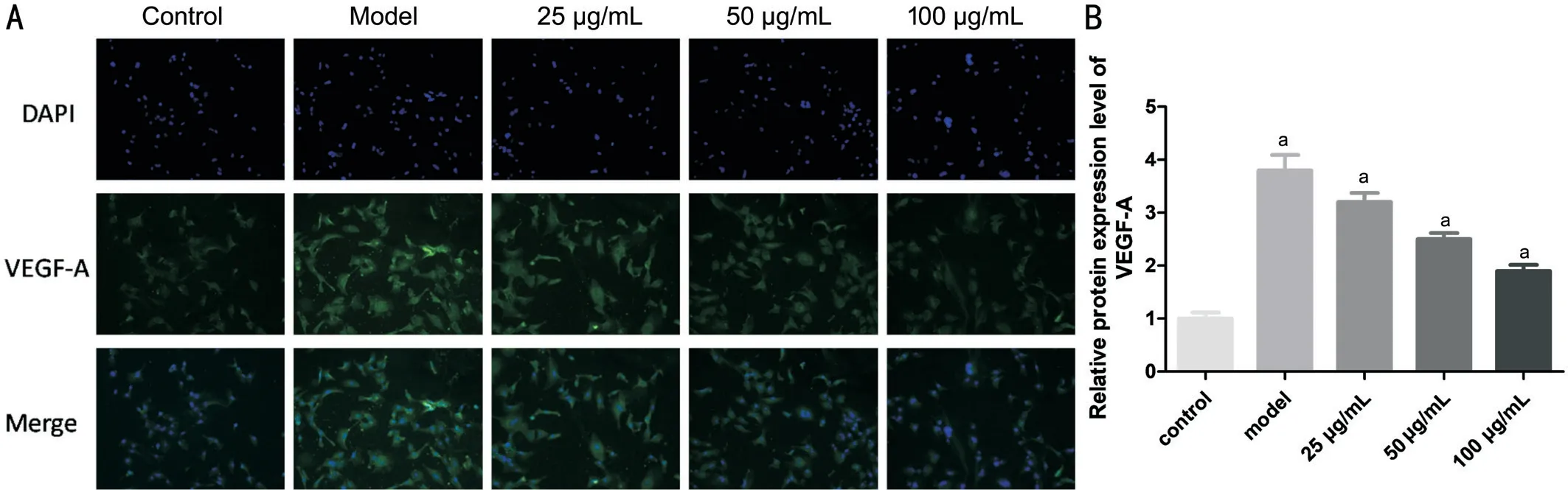
Figure 5 VEGF-A expression was observed under a fluorescence microscope after immunostaining DAPI was used for staining the nuclei.A: Immunohistochemistry showed positive staining of VEGF-A in the hUCMSC exosome-treated HRECs compared to that of the model group.B: Relative expression of the VEGF-A protein in the HRECs in each group. aP<0.05 compared with control.
After exosomes are extracted, NTA should be performed to confirm that exosomes with a diameter between 50-150 nm are obtained rather than nanoscale components with larger diameters, such as cell vesicles. NTA involves tracking the scattering of a single particle and performance of mathematical calculations based on the concentration of exosomes and the hydrodynamic diameter. Methods to identify the composition of exosomes include Western blot analysis, trypsin digestion,flow cytometry, mass spectrometry, and ELISAs[22]. Since all exosomes are derived from endosomes, they contain the same fusion proteins and membrane transporters (Annexins, flotillin,GTPases), tetraspanins (CD9, CD82, CD81 and CD63),heat shock proteins (such as Hsp20, Hsp60, and Hsp70),proteins involved in the formation of MVB (TSG101, Alix),phospholipase and lipid-related membrane proteins[23].
In this study, a certain number of samples were obtained by the most widely used low-temperature ultracentrifugation method,the membrane structure was observed by (transmission electron microscopy) TEM, the diameter of the sample was detected by NTA, and the sample was verified as exosomes through characterization and the particle diameter. Two kinds of tetraspanin, CD9 and CD63, and the heat shock protein HSP70 were selected as exosome-specific proteins for detection, and GRP94, which is not located in the plasma membrane or endosomes, was selected as a negative control.GRP94 exists in some extracellular vesicles but is not enriched in vesicles with an inner diameter of approximately <200 nm in plasma membrane endosome-derived vesicles. In summary,we confirmed that the samples we extracted in the experiment were hUCMSC-derived exosomes.
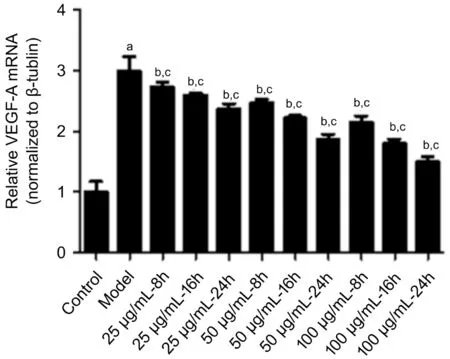
Figure 6 The VEGF-A mRNA levels were found to be markedly decreased in the hUCMSC-derived exosome-treated high glucoseinduced HRECs at 8, 16, and 24h compared to those of the model group mRNA levels of VEGF-A decreased consistently over time and with increasing concentrations of the hUCMSC-derived exosomes.aP<0.05 compared with the control; bP<0.05 comparison of the model group and the treatment groups at the same concentration; cP<0.05 comparison of the model group and the treatment groups at the same time.
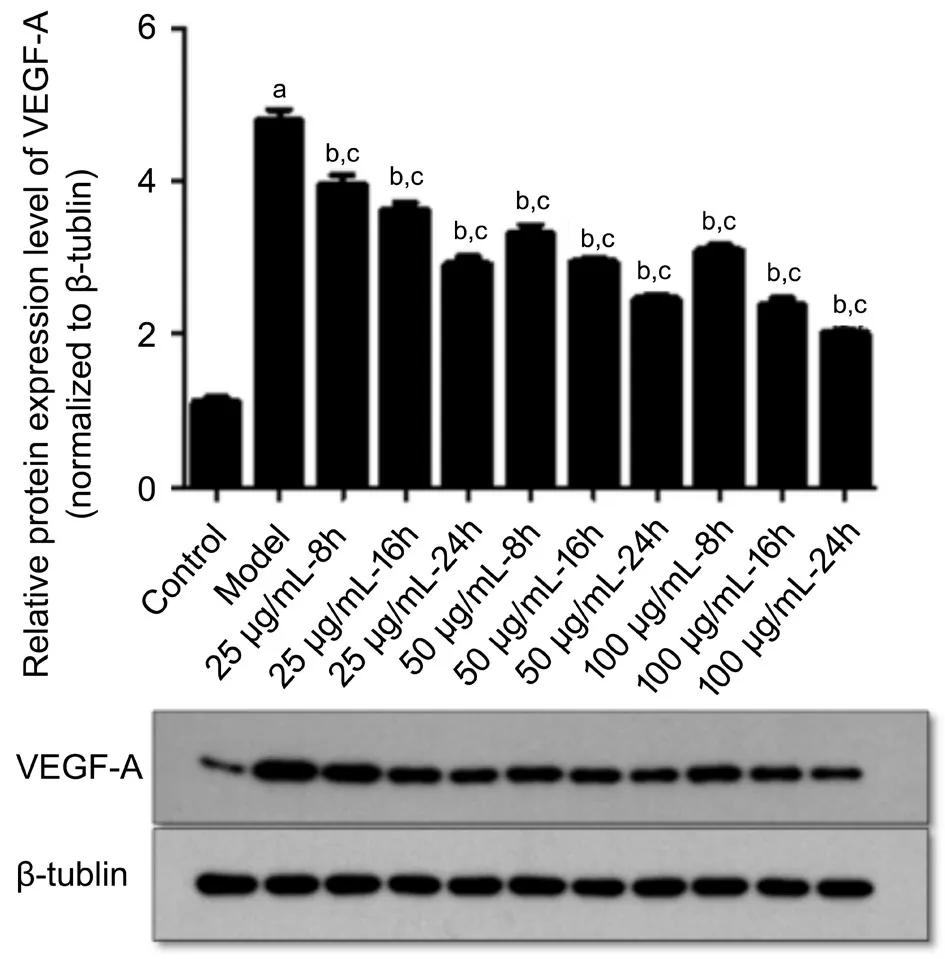
Figure 7 The protein levels of VEGF-A decreased consistently over time and with increasing concentrations of the hUCMSCderived exosomes aP<0.05 compared with the control; bP<0.05 comparison of the model group and the treatment groups at the same concentration; cP<0.05 comparison of the model group and the treatment groups at the same time.
MSC-derived exosomes play a critical role in angiogenesis.According to the bioinformatics analysis conducted by Onoet al[24], angiogenesis is related to genes targeted by miRNAs in MSC-derived exosomes. Zhuet al[25]reported that MSC-derived exosomes can increase the expression of VEGF. Adipose-derived mesenchymal stem cell (ASC)-derived exosomes are essential for endothelial cell (HMEC)angiogenesisin vivoandin vitro. Platelet growth factor (PDGF)enhances this activity by stimulating the release of exosomes and microvesicles from ASCs[26]. Under hypoxic conditions,MSC-derived exosomes deliver mRNAs and miRNAs to targeted cells and promote the proliferation of endothelial cells, thus leading to angiogenesis and enhancing blood flow recovery and capillary network formation[27]. In contrast,studies by Pakravanet al[28]and Leeet al[29]both suggested that MSC-derived exosomes significantly reduced the expression and secretion of VEGF in a dose-dependent manner, which was consistent with the results we obtained. In our study,hUCMSC-derived exosomes were cocultured with HRECs induced by high glucose, and the hUCMSC-derived exosomes had an inhibitory effect on angiogenesis and downregulated the level of VEGF-A in HRECs, verifying the anti-VEGF effect of the MSC-derived exosomes on neovascular retinal diseases by cell experiments. Recent research has revealed that the MSC-derived exosomes can inhibit the expression of hypoxia-induced mitotic factor (HIMF) and thus inhibit cell proliferation, suggesting that MSC-derived exosomes play a vital role in anti-vascular remodelling[30-32].
There are no detailed studies on the biological effect of huMSC-derived exosomes on the expression of VEGF-A.Some studies have reported that MSC-derived exosomes regulate VEGF-A through miRNAs. For instance, miR-125a[33]and miR-30b[34]can promote the formation of human umbilical vein endothelial cells by inhibiting the angiogenesis inhibitor delta-like ligand 4 (DLL4). Interestingly, MSC-derived exosomes also contain antiangiogenic miRNAs, such as miR-16[30]and miR-100[28], and inhibit angiogenesis by targeting VEGF in blood vessels and breast cancer cells in the tumour microenvironment. Zhanget al[35]found that miR-320a loaded with exosomes can downregulate HIF-1α expression,resulting in a decrease in the expression of VEGF-Ain vitro. It has been revealed that microglia-derived exosomes can inhibit hypoxia-induced photoreceptor apoptosis in the (oxygeninduced retinopathy) OIR model through miR-24-3p, and it has also been proven that exosomes can inhibit the expression of proangiogenic factors in the photoreceptorin vitroby downregulating VEGF and TGF-β expression by inhibiting the phosphorylation of Akt and ERK[36].
In summary, this study focused on MSC-derived exosomes that may have anti-VEGF biological activity. Although the mechanism of action is not clear, the various characteristics of exosomes, which possess the biological activity of MSCs but do not cause the various live cell transplantation-induced complications, have been incorporated. Exosomes have enzyme-centred characteristics that can adjust their biological activity according to the environment, reducing the risk of overdose or underdose. Based on these findings along with their nanometre-scale diameter, easy preparation and production, and many other advantages, hUCMSC-derived exosomes are safer and more effective than MSCs, and they could be used as a new and effective treatment tool to protect the retina.
ACKNOWLEDGEMENTS
Foundation:Supported by Science and Technology Fund of Tianjin Eye Hospital (No.YKYB1905).
Conflicts of Interest:He GH, None; Ma YX, None; Dong M,None; Chen S, None; Wang YC, None; Gao X, None; Wu B,
None; Wang J, None; Wang JH, None.
 International Journal of Ophthalmology2021年12期
International Journal of Ophthalmology2021年12期
- International Journal of Ophthalmology的其它文章
- Comparison of trifocal toric and bifocal toric intraocular lens implantation in patients with cataract and high corneal astigmatism
- Comparison of perioperative parameters in one-handed rotational phacoemulsification versus conventional phacoemulsification and femtosecond laser-assisted cataract surgery
- Decreased retinal microvasculature densities in pterygium
- Fourier analysis of corneal Scheimpflug imaging: clinical use in keratoconus
- Establishment of a prediction tool for ocular trauma patients with machine learning algorithm
- Role of orthoptics and scoring system for orbital floor blowout fracture: surgical or conservative treatment
