Relationship between miR-7-5p expression and 125 I seed implantation efficacy in pancreatic cancer and functional analysis of target genes*
Tingting Hao ,Chaoqi Wang (Co-first author),Yingjie Song (Co-first author),Wanyan Wu ,Xuetao Li ,Tao Fan (✉)
1 Department of Oncology,The People's Hospital of China Three Gorges University,The First People's Hospital of Yichang,Yichang 443000,China
2 Department of Urinary Surgery,Affiliated Hospital of Inner Mongolia University for the Nationalities,Tongliao 028007,China
3 General Surgery,The People's Hospital of China Three Gorges University,The First People's Hospital of Yichang,Yichang 443000,China
Abstract Objective The aim of this study was to investigate the relationship between miR-7-5p expression and intertissue-125I irradiation sensitivity in pancreatic cancer tissues and to analyze the function of target genes.Methods Thirty-seven patients with unresectable pancreatic ductal adenocarcinoma (PDAC) treated with radioactive 125I seed implantation were enrolled.RT-PCR was used to detect the expression level of miR-7-5p in cancer tissues and analyze the relationship between miR-7-5p expression and 125I radiation sensitivity.Bioinformatic software and online tools were used to predict the miR-7-5p target genes and analyze their functional annotation and pathway enrichment.Results Radioactive 125I seed implantation was followed up for 2 months.The objective response rate of the miR-7-5p high expression group was 65.0% (13/20),whereas the objective response rate of the miR-7-5p low expression group was 5.88% (1/17),and the difference between the two groups was statistically significant (χ2=13.654,P <0.001).A total of 187 target genes were predicted using three databases.GO functional annotation showed that target genes were mainly involved in cellular response to insulin stimulus,regulation of gene expression by genetic imprinting,cytosol,peptidyl-serine phosphorylation,bHLH transcription factor binding,cargo loading into vesicles,cellular response to epinephrine stimulus,and nucleoplasm.KEGG pathway enrichment analysis showed that target genes were mainly involved in the ErbB signaling pathway,HIF-1 signaling pathway,axon guidance,longevity regulatory pathway,endocrine resistance,glioma,choline metabolism in cancer,and EGFR tyrosine kinase inhibitor drug resistance.Molecular complex detection analysis by Cytoscape revealed that PIGH,RAF1,EGFR,NXT2,PIK3CD,PIK3R3,ERBB4,TRMT13,and C5orf22 were the key modules of miR-7-5p target gene clustering.Conclusion The expression of miR-7-5p in pancreatic cancer tissues positively correlated with the radiosensitivity of 125I seeds.Via targeted gene regulation,miR-7-5p acts on the network of multiple signaling pathways in PDAC and participates in its occurrence and development.Thus,miR-7-5p may become a predictive index of 125I seed implantation therapy sensitivity in PDAC patients.
Key words: miR-7-5p;pancreatic cancer;125I;radioactive seed implantation
The relationship between miRNAs and tumors is similar to that of tumor suppressor genes and cancer activators.The miRNAs are directly or indirectly involved in regulating the expression of one or more target genes.Abnormal expression of miRNAs plays an important role in the occurrence and development of tumors as well as drug efficacy evaluation[1–3].Therefore,miRNAs are increasingly being considered potential biomarkers for disease diagnosis,curative effect judgment,and prognosis evaluation[4].We studied the effect of125I seed implantation on unresectable pancreatic ductal adenocarcinoma(PDAC) and the expression level of miR-7-5p in cancer tissue,to explore the relationship between the expression of miR-7-5p and the irradiation sensitivity of radioactive125I seeds.Bioinformatic software and online tools were used to predict miR-7-5p target genes,perform their functional annotation and pathway enrichment analysis,and analyze the key modules of the target gene network.
Materials and methods
This study was conducted at the People’s Hospital of China Three Gorges University,China,from January 2017 to March 2019.The research was conducted after approval from the Committee on Human Research and Ethics.Thirty-seven patients with unresectable PDAC undergoing radioactive125I seed implantation were enrolled in the study.Complete follow-up data were collected.The125I isotope,activity 0.5–0.9 mCi,half-life 59.43 days,gross tumor volume (GTV) prescription dose 115–130 Gy (at least D90 reached the prescription dose).
The expression of miR-7-5p in pancreatic carcinoma was detected by real-time PCR.The total mRNA kit from Takara Company was used to extract total cellular RNA.cDNA was prepared as described in the instructions of the Prime Script®RT Kit with gDNA Eraser (Perfect Real Time) reverse transcription kit.Real-time PCR amplification was conducted using cDNA as a template and U6 as an internal reference.The miR-7-5p upstream primer was 5’-CCACGTTGGAAGACTAGTGATTT-3’,and the downstream primer was 5’-TATGGTTGTTCT GCTCTCTGTCTC-3’.U6 upstream primer:5’-CTCGC TTCGGCAGCACA-3’;downstream primer:5’-AACGC TTCACGAATTTGC GT-3’.The reaction mixture consisted of 10.0 μL of 2 × SYBR Premix Ex TaqTMII,0.8 μL of each of the upstream and downstream primers,2.0 μL of cDNA,0.4 μL of ROX reference dye (50 ×),and DEPC water to a total of 20.0 μL.The reaction conditions were as follows:denaturation at 95°C for 30 s,annealing at 62°C for 40 s,repeating the above steps for 40 cycles,and finally a hold at 4°C for preservation.After the end of PCR amplification,the analyzer displayed the standard curve,amplification curve,and melting curve.The amplification factor was calculated using the 2-ΔΔCtmethod.
Three online tools,TargetScan,miRDB,and miRWalk,were used to predict the target genes of hsa-miR-7-5p,and to obtain a more accurate target gene by creating a Venny map (https:// bioinfogp.cnb.csic.es/tools/venny/index.html) and taking the intersection.An online platform (https:// cloud.oebiotech.cn/task/detail/arrayenrichment/) was used to perform Gene Ontology(GO) functional annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) was used to perform pathway enrichment analysis.The online tool STRING(https://string-db.org/) and miRNet 2.0 database (www.mirnet.ca/Faces/home.xhtml) were used to obtain the miRNA-Gene network graph and matrix list,which was then imported using Cytoscape 3.6.1 software.The Molecular Complex Detection (MCODE) plugin was used for module clustering analysis to detect the potential functional modules in the network.In the MCODE process,the cut-off value of a degree was set to 2,and the cut-off value of the node score was set to 0.2.
SPSS version 20.0,was used for statistical analysis of the data.Cancerous tissue miR-7-5p relative expression was measured using the 2-ΔΔCtvalue measure,divided into high-and low-level groups.Measurement data are expressed as mean ± SD.An independent samplet-test was used for comparison between the two groups.AllPvalues were bidirectional andP<0.05 was considered to be significant.
Results
Among the 37 patients who received CT-guided radioactive125I seed inter-tissue implantation therapy,20 patients had high miR-7-5p expression (miR-7-5p High)and 17 patients had low miR-7-5p expression.Thirtyseven patients were treated with inter-tissue implantation of radioactive125I seeds.After 2 months of follow-up,14 patients achieved PR,23 achieved SD,and no patients achieved CR or PD.The objective response rate of the miR-7-5p high group was 65.0% (13/20) whereas that of the miR-7-5p low group was 5.88% (1/17),and there was a significant difference between the two groups (χ2=10.347,P=0.001;Fig.1).The results showed that the expression of miR-7-5p positively correlated with the irradiation sensitivity to radioactive125I seeds.Patients with high expression of miR-7-5p were relatively sensitive to radiotherapy with high objective efficiency,while patients with low expression were relatively insensitive to radiotherapy with low objective efficiency.
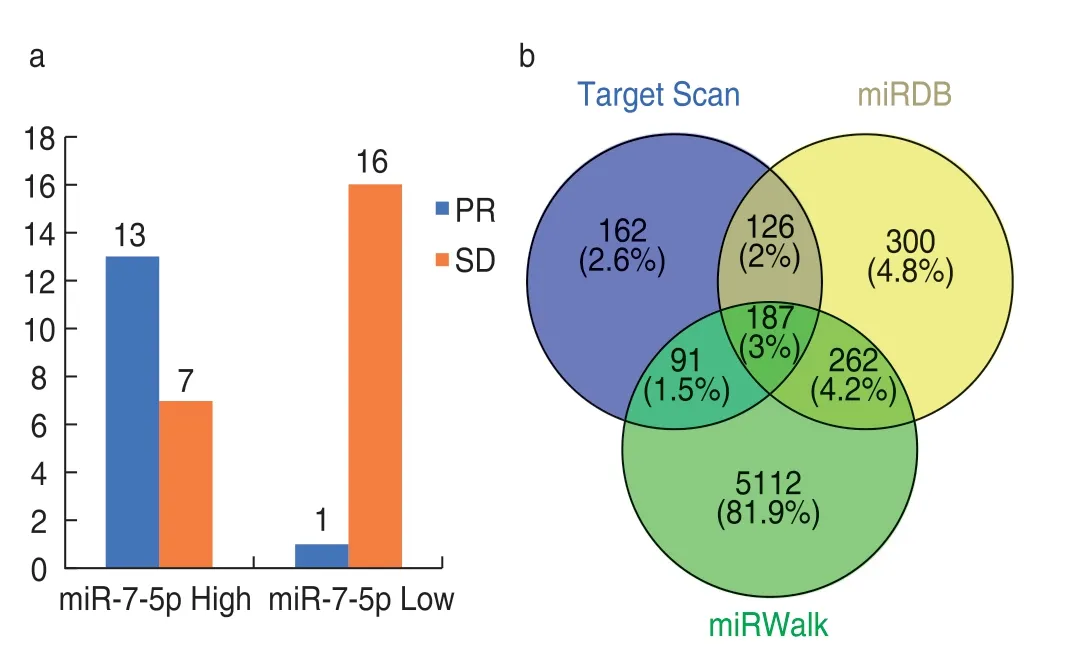
Fig.1 (a) Relationship between miR-7-5p expression level and therapeutic response;(b) Venn diagrams of TargetScan,miRDB,and miRWalk predicting miRNA-7-5p target genes
The online tools TargetScan,miRDB,and miRWalk were used to predict target genes of miR-7-5p and a total of 187 target genes were identified by all three tools (Fig.1).Through online tools,mirDIP 4.1 (http://ophid.utoronto.ca/ mirDIP/index confirm JSP) filtering.The lowest reliability of setting parameters was selected as very high,and 25 prediction software packages,such as filter source RNA22,Mirtar,RNAHybrid,Mircode,MBStar,TargeRank,Microrna.org,Pita,TargetScan,PicTar,and MirBase were selected,and the frequency of occurrence of target genes was at least 1.The results showed that the 187 intersection target genes were all target genes of very high credibility.
There were 135 biological process (BP) annotated genes,146 cellular component (CC) annotated genes,and 129 molecular function (MF) annotated genes.A bar chart for the top 10 most significant functions of BP,CC,and MF genes identified through the GO enrichment analysis is shown in Fig.2.
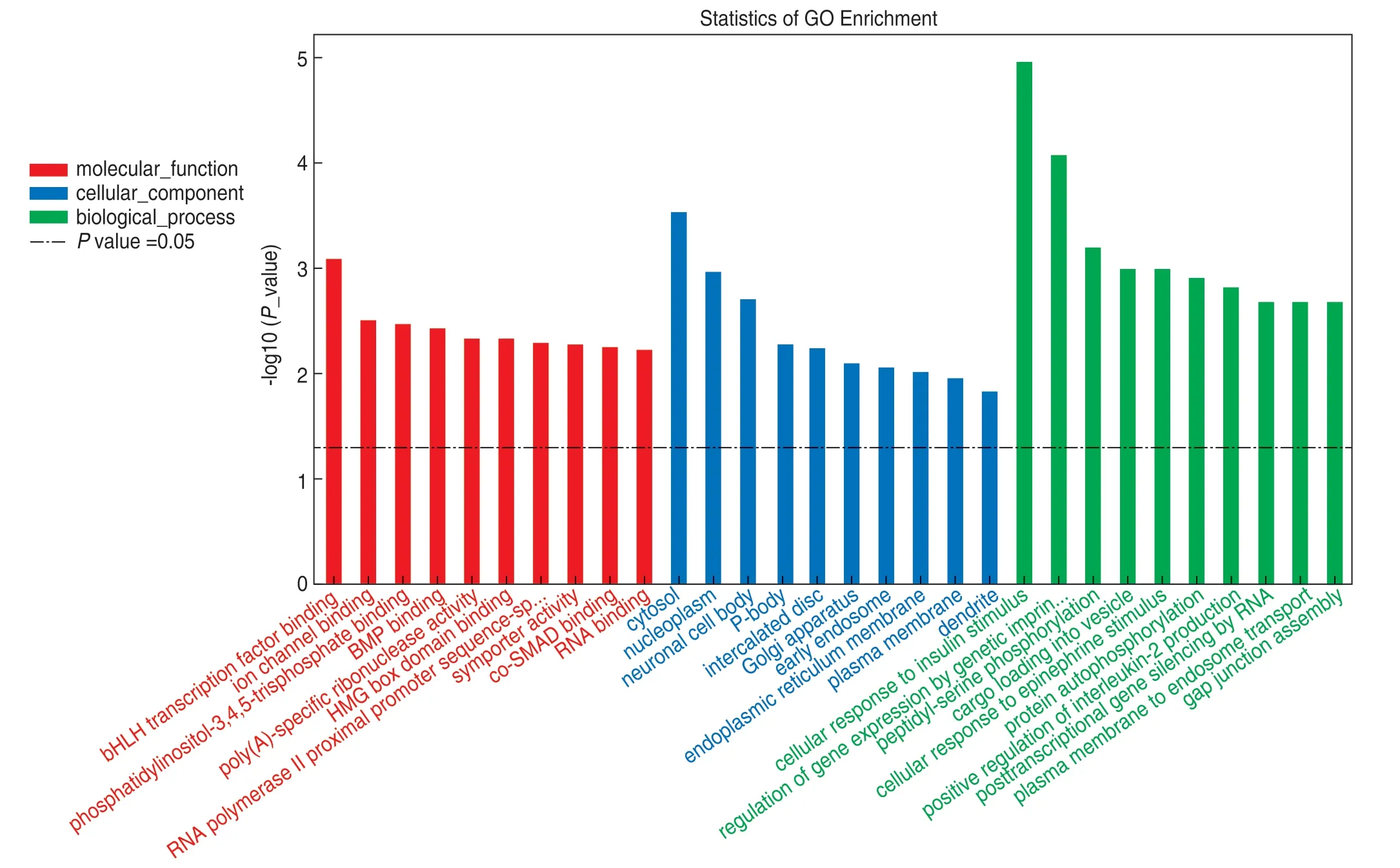
Fig.2 GO bar graph.Y-axis is -log10 (P value).The higher the bar graph height,the greater the significance
Through the BP,CC,and MF functional annotation of GO,we found that target genes were mainly involved in cellular response to insulin stimulus,regulation of gene expression by genetic imprinting,cytosol,peptidylserine phosphorylation,bHLH transcription factor binding,cargo loading into vesicles,cellular response to epinephrine stimulus,and nucleoplasm.KEGG pathway enrichment analysis showed that target genes were mainly involved in the ErbB signaling pathway,HIF-1 signaling pathway,axon guidance,longevity regulatory pathway,multiple species,endocrine resistance,glioma,choline metabolism in cancer,and EGFR tyrosine kinase inhibitor drug resistance (Fig.3).
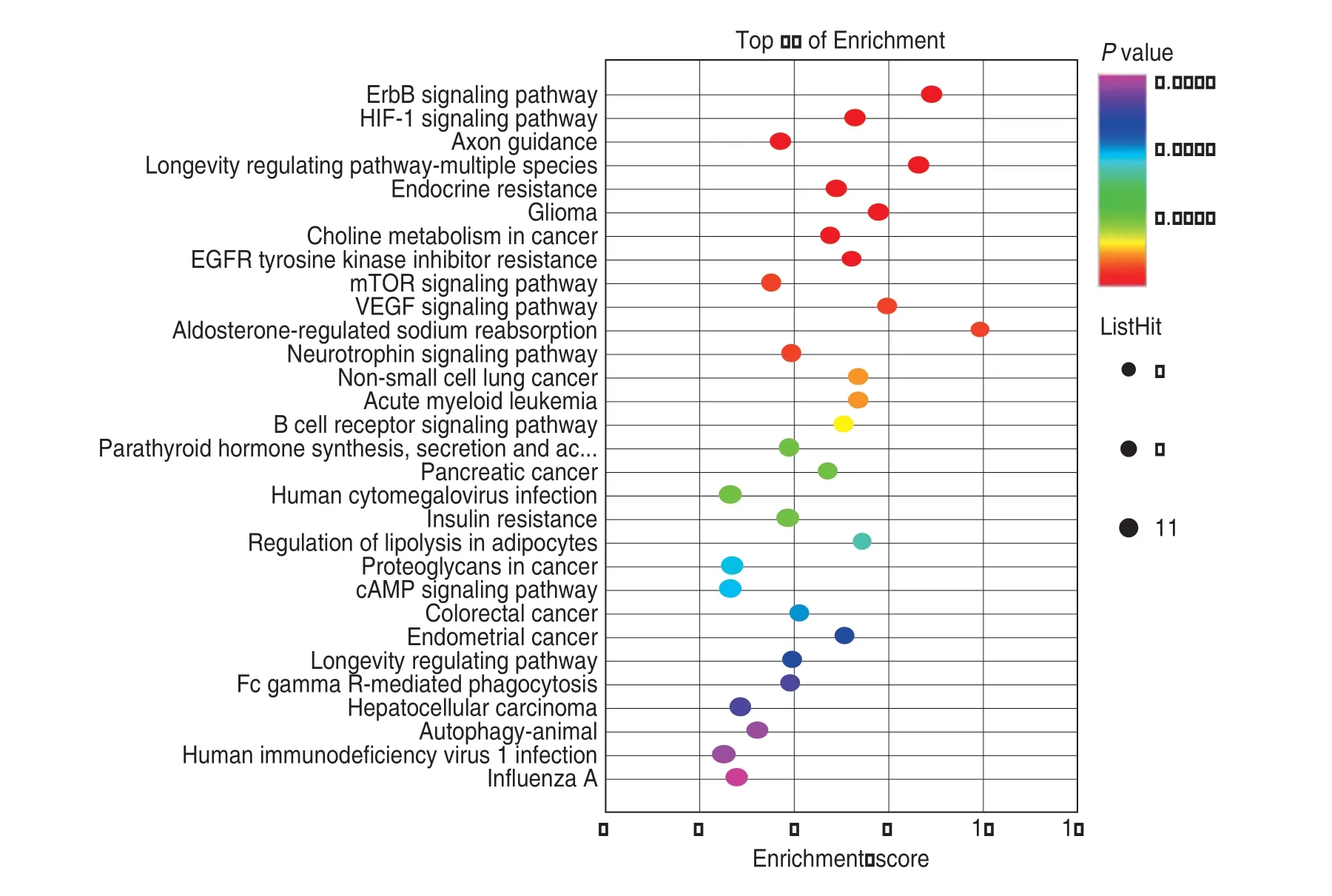
Fig.3 KEGG bubble chart:the y-axis corresponds to the KEGG entries,the x-axis corresponds to the enrichment score,the size of the point corresponds to the number of intersection genes in the KEGG entries,and the smaller the P value of KEGG enrichment,the greater the significance
Seven potential functional modules in the network were identified using module clustering (Table 1).PIGH,RAF1,EGFR,NXT2,PIK3CD,PIK3R3,ERBB4,TRMT13,and C5orf22 were the key modules in the hsa-miR-7-5p target gene cluster.

Table 1 Seven functional modules of MCODE module cluster analysis
Discussion
MiR-7-5p has been reported to play an anti-tumor role in human colorectal cancer[5],NSCLC[6],nasopharyngeal carcinoma[7],gastric cancer[8],hepatocellular carcinoma[9],cervical cancer[10],breast cancer[11],small cell lung cancer[12],and other tumors.However,its expression and function are sometimes contradictory.The expression of miR-7-5p is significantly lower in lung cancer tissues.In vivoandin vitrostudies have shown that miR-7-5p targeting NOVA2 inhibits the proliferation,migration,and invasion of lung cancer cells[6].In esophageal cancer tissues,miR-7-5p is highly expressed compared to para-cancerous tissues and is negatively correlated with the survival rate of patients with esophageal cancer.The downregulation of miR-7-5pin vivocan inhibit the occurrence and development of esophageal cancer by inactivating the MAPK signaling pathway[13].Nevertheless,the high expression level of miR-7-5p can significantly inhibit the growth,migration,and colony formation of human hepatocellular carcinoma cells[14].
In addition,miR-7 modulates the chemoradiotherapy sensitivity of some tumors.In terms of chemotherapy resistance,Laiet al.revealed that miR-7 is downregulated in doxorubicin-resistant small cell lung cancer cells,and miR-7 enhancement inhibits doxorubicin-induced homologous recombination repair by inhibiting the expression of Rad51 and BRCA1,thereby improving the sensitivity of doxorubicin-resistant small cell lung cancer cells to doxorubicin[12].In temozolomide (TMZ)-resistant gliomas,miR-7 can increase the sensitivity of cells to TMZ by downregulating YY1 and hindering the stemness of TMZ-resistant glioblastoma cells[15].In terms of radiation resistance,studies have found that high expression of miR-7-5p can promote radiation resistance in cervical cancer and liver cancer cells after radiation (5 Gy and 10 Gy)[16].However,other studies have found that miR-7-5p inhibitor can significantly enhance the radiation resistance of nasopharyngeal carcinoma cells by promoting the proliferation,migration,and invasion of NPC cells,inhibiting cell apoptosis,and improving the sensitivity of nasopharyngeal carcinoma cells to radiotherapy[17].The role of miR-7-5p in the radiosensitivity of human pancreatic cancer has not yet been studied.In this study,we found that the expression of miR-7-5 was positively correlated with the radiosensitivity of pancreatic cancer.Thirty-seven patients with unresectable pancreatic cancer were treated with125I seeds under the guidance of spiral CT.After 2 months of follow-up,14 patients achieved PR,23 patients achieved SD,and no patients achieved CR or PD.The objective response rate (ORR)of the miR-7-5p high group was 65.0% (13/20) whereas the ORR of the miR-7-5p group was 5.88% (1/17),and there was a significant difference between the two groups(χ2=13.654,P<0.001).The results showed that the high expression of miR-7-5p was relatively sensitive to radiotherapy with a high objective effective rate,while the low expression of miR-7-5p was relatively insensitive to radiotherapy with a low objective effective rate.
The tumorigenicity and tumor inhibition of miRNA are achieved by binding to the 3’ noncoding region of the corresponding target gene to degrade or inhibit the translation of the target gene miRNA[18].Because of the diversity of miRNA target genes in type and quantity,the same miRNA may regulate multiple target genes simultaneously,and the same gene may be precisely regulated by multiple miRNAs at the same time.The regulatory mechanisms of target genes are complex.Therefore,it is very important to accurately predict miRNA target genes and correctly understand the regulation mode between miRNA and target genes[19].
Currently,bioinformatics is an important method for predicting miRNA target genes[20]and has the advantage of being able to handle a large amount of data and simple operation.To improve the accuracy of target gene prediction and reduce false results,the results of multiple database prediction are intersected.In this study,187 miRNA-7-5p target genes were predicted and screened using TargetScan,miRDB,and miRWalk.BP,CC,and MF enrichment analysis of GO showed that target genes were mainly involved in cellular response to insulin stimulus,regulation of gene expression by genetic imprinting,cytosol,peptidyl-serine phosphorylation,bHLH transcription factor binding,cargo loading into vesicles,cellular response to epinephrine stimulus,and nucleoplasm.KEGG pathway enrichment analysis showed that target genes were mainly involved in the ErbB signaling pathway,HIF-1 signaling pathway,axon guidance,longevity regulatory pathway,multiple species,endocrine resistance,glioma,choline metabolism in cancer,and EGFR tyrosine kinase inhibitor drug resistance.Using the mirnet2.0 database and Cytoscape 3.6.1 software,we found that PIGH,RAF1,EGFR,NXT2,PIK3CD,PIK3R3,ERBB4,TRMT13,and C5ORF22 were the key modules in the cluster of hsa-miR-7-5p target gene networks.However,the rationality of miR-7-5p and its target gene as a biomarker of radiotherapy sensitivity in pancreatic cancer requires further study.
Conclusion
This study found that age,sex of and tumor marker level,tumor location,and tumor size in patients with pancreatic cancer were not the main factors affecting the efficacy of radioactive125I particles.However,the expression of miR-7-5p in pancreatic cancer tissues positively correlated with the radiosensitivity of125I seeds.Through targeted gene regulation,miR-7-5p acts on the network of multiple signaling pathways of PDAC and participates in its occurrence and development and is thus expected to become a predictive index of125I seed implantation therapy sensitivity in patients with PDAC.
Conflicts of interest
The authors indicated no potential conflicts of interest.
 Oncology and Translational Medicine2021年4期
Oncology and Translational Medicine2021年4期
- Oncology and Translational Medicine的其它文章
- The expression of vascular endothelial growth factor (VEGF)/ endostatin (ES) and VEGF receptor 2(VEGFR2)/ES is associated with NSCLC prognosis *
- Relationship between molecular changes in epidermal growth factor receptor (EGFR)and anaplastic lymphoma kinase (ALK)mutations in lung adenocarcinoma *
- Efficacy and adverse reactions of apatinib as secondline or later-line treatment in advanced lung cancer
- Enhancing the treatment effects of tumor cell purified autogenous heat shock protein 70-peptide complexes on HER-3-overexpressing breast cancer *
- The clinical efficacy of percutaneous ethanol-lipiodol injection (PEI) combined with high-intensity focused ultrasound (HIFU) for small hepatocellularcarcinoma in special or high-risk locations*
- Mechanism of tumor synthetic lethal-related targets
