7SK truncation at 128-179 nt suppresses embryonic stem cell proliferation in vitro by downregulating CDC6
CHEN Rui ,ZHANG Yurong ,CHEN Peng ,PANG Yixin ,LI Hongbao ,CHEN Ziwei ,ZHANG Xiaoyong,ZHANG Hongyi,LI Wujun
1First Affiliated Hospital of Xi'an Medical University,Xi'an 710077,China;2Institute of Basic Medical Science,Xi'an Medical University,Xi'an 710021,China;3Second Affiliated Hospital of Xi'an Jiaotong University,Xi'an 710004,China;4Department of Physiology and Pathophysiology,Xi'an Jiaotong University School of Basic Medical Sciences,Xi'an 710061,China;5School of Clinical Medicine,Xi'an Medical University,Xi'an 710021,China
Abstract: Objective To explore the role of small nuclear noncoding RNA 7SK in embryonic stem cell(ESCs)proliferation and the value of 7SK as a target for early diagnosis and treatment for primordial dwarfism(PD).Methods ESC line R1 was transfected with the CRISPR/Cas9 system,and sequencing of the PCR product and glycerol gradient analysis were performed to identify novel 7SK deletion mutations.A lentivirus system was used to knock down cyclin-dependent kinase 9 (CDK9) in clones with 7SK deletion mutations,and the effect of CDK9 knockdown on the protein level of cell division cycle 6 (CDC6) was analyzed with Western blotting.Results We identified a novel deletion mutation of 7SK at 128-179 nt in the ESCs,which resulted in deficiency of cell proliferation.7SK truncation at 128-179 nt significantly reduced the protein expressions of La-related protein 7(LARP7)and CDC6.Conclusions 7SK truncation at 128-179 nt can significantly impair proliferation of ESCs by downregulating CDC6.7SK is a key regulator of proliferation and mediates the growth of ESCs through a mechanism dependent on CDK9 activity,suggesting the value of 7SK truncation at 128-179 nt as a potential target for early diagnosis and treatment of PD.
Keywords:7SK truncation;embryonic stem cell;proliferation;cyclin-dependent kinase 9;cell division cycle 6
INTRODUCTION
Primordial dwarfism(PD),characterized by global growth failure both during embryogenesis and postnatally,results in smaller sizes of most of the body parts while maintaining their normal anthropometric proportions in an affected individual[1].Mutations in U4atac small nuclear RNA (snRNA,a component of the minor spliceosome),among other genes,have been linked to PD,but how those genes contribute to PD remains poorly understood[1].
Studies have indicated the important role of disturbances of embryonic stem cell(ESCs)development in the pathogenesis of PD.Pluripotent ESCs are capable of generating all the cell types seen in an adult organism[2].During the early stages of development,ESCs maintain a delicate balance between self-renewal and differentiation,and disruption of this balance can lead to premature ESC differentiation and causes a spectrum of developmental diseases including PD[3,4].Even in the primed state,the ESCs still maintain their self-renewal ability and pluripotency,which is controlled by the same set of core pluripotency transcription factors as in their naive state[5].
A previous study suggested that La-related protein 7(LARP7) mutation could cause primordial dwarfism(PD)[6].In animal models,a loss-of-function mutation of LARP7 was found to result in a smaller size of the animal[7].Additionally,most of the PD-related genes are involved in DNA replication and damage response pathway[8].Among these genes,7SK encodes a highly conserved small nuclear noncoding RNA consisting of 332 nucleotides[9],which interacts with LARP7viaits short 3'-terminal and binds to hexamethylene bisacetamide-inducible proteins 1 (HEXIM1) and 2(HEXIM2) and the methylphosphate capping enzyme(MePCE)viaits long 5'-terminal[10].So far the regulatory role of 7SK in cell proliferation remains unclear,but this non-coding RNA is found to negatively regulate positive transcription elongation factor b(P-TEFb)[11,12],a complex composed of cyclin-dependent kinase 9 (CDK9) and cyclin T(cyclin T1 and T2)essential for phosphorylation of RNA polymerase II to promote RNA synthesis[13].These observations suggest the essential role of 7SK in regulating cell proliferation.
Here we show that nucleotide deletion of 7SK gene at 128-179 nt by a CRISPR/Cas9 system causes deficiency in proliferation of ESCs without affecting the self-renewal ability of the cells;this deletion also reduces the protein levels of LARP7 and cell division cycle 6(CDC6).Our findings strongly indicate that 7SK is a key regulator of ESC proliferation and mediates cell growth through a mechanism dependent on CDK9 activity.We hypothesize that the regulation of deletion of 7SK gene at 128-179 nt and the expression of CDC6 are essential to maintain a sufficient number of undifferentiated ESCs and hence the normal size of an organism.
METHODS
Plasmids
CRISPR/Cas9 targeting 7SK was cloned into a px459 vector (Addgene) following the protocols described by Ran et al[14].The positive clones with the target gene knockout were determined by Western blotting and sequencing.CDK9 knockdown plasmid was cloned into the pLKO.1 vector (Addgene).The sequences for CDK9 knockdown are listed below:
sh-Cdk9-1,
CCGGCGTCTTAGTCAAGTTCACGTTCTCGAGAACGT GAACTTGACTAAGACGTTTTTG;
sh-Cdk9-2,
CCGGCATGAGAATGTGGTGAACCTACTCGAGTAGG TTCACCACATTCTCATGTTTTTG;
shCdk9-3,
CGGGTACGAGAAACTTGCCAAGATCTCGAGATCTT GGCAAGTTTCTCGTACTTTTTG).The positive clones with the target knockout were identified by sequencing(Tsing Ke Biological Technology) and glycerol gradient analysis.
CDC6 overexpression plasmid was cloned into a pLVX-IRES-ZGreen vector containing the restriction endonuclease siteEcoR1/BamH1 following the manufacturer's instructions(Thermo Fisher).
Antibodies
The following commercial antibodies were used:anti-Nanog(Santa Cruz,1∶1000 dilution),CDC6(Santa Cruz,1∶1000 dilution),Cyclin T1(Santa Cruz,1∶1000 dilution)and CDK9 (Santa Cruz,1∶2000 dilution),anti-GAPDH(KangChen,1∶20 000 dilution) and MCM2 (KangChen,1∶3000 dilution),and anti-LARP7 (Abmart,1∶5000 dilution).
Cell culture and treatment
The embryonic stem cell line (R1) was purchased from American Type Culture Collection (ATCC) and cultured following the ATCC guidelines.Specifically,the cells were cultured in Dulbecco's Modified Eagle's Medium(DMEM) containing 15% fetal bovine serum (Gemini Bio-Products) and supplemented with 1×103U/mL leukemia inhibitory factor (LIF;Millipore),1 mmol/L sodium pyruvate,100 mmol/L 2-mercaptoethanol,and 100 mmol/L nonessential amino acids (Invitrogen).For knockout experiment,the cells were transfected with the CRISPR/Cas9 constructsvialipofectamine 2000(Invitrogen).After 24 h,the cell culture was treated with 1.5 μg/mL puromycin to remove the untransfected cells.
Cell transfection
The lentivirus was transferred into psPAX2,pMD2.G,and Scr,shCDK9 plasmids and packaged in 293T cells.Two days after transfection,centrifuged at 1500×gfor 5 min,and then filtered with a 0.45 filter.ESCs(R1)were transduced with the lentivirus carrying pLKO.1 shRNA construct for two days,and were then seeded into 6-well plates (1×105cells per well) containing growth medium supplemented with 1.5 mg/mL puromycin (Sigma) for culture for another 3 days before Western blot analysis.
Glycerol gradient analysis
Glycerol gradient analysis was carried out following the protocols described previously[4].Briefly,R1 cells were detached,centrifuged at 2000 r/min,and lysed in the cell lysis buffer (containing 150 mmol/L NaCl,2 mmol/L MgCl2,10 mmol/L HEPES,1 mmol/L EDTA,1 mmol/L DTT,1% PMSF,EDTA-free protease inhibitor [Roche],and 0.5%Nonidet P-40)for 10 min on ice.The cell lysate was centrifuged for 15 min at 13 000 r/min,and the supernatant was loaded onto 5%-45% glycerol gradients in the cell lysis buffer free of Nonidet P-40.The gradients were run at 49 500 r/min for 16 h in a Beckman MLS-50 rotor and then fractionated.
Protein extraction and Western blotting
The cells were transferred to a 1.5 mL Eppendorf tube containing pre-cooled extraction buffer (50 mmol/L HEPES [pH 7.5],140 mmol/L NaCl,1 mmol/L EDTA[pH 8.0],1% Trition X-100,0.1% sodium deoxycholate,0.1% SDS,protein inhibitor [Roche],and 0.1% PMSF).The cell lysate was separated by 10% SDS-PAGE,and the protein fractions were electrotransferred to a PVDF membrane (Millipore,Bedford,MA,US),which was blocked and incubated with the primary antibodies overnight at 4 ℃.After incubation with horseradish peroxidase (HRP)-conjugated secondary antibodies for 1 h at room temperature,the protein blots were visualized with electrochemiluminescence (ECL) reagents by a chemiluminescence imaging system(Bio-Rad).
Growth curve
The wild-type R1 (R1-WT) cells and those with 7SK truncation were seeded in triplicates in 6-well plates.After digestion with trypsin on days 2,4,and 6,the cells were counted and statistically analyzed.
Statistical analysis
The protein expressions determined by Western blotting were quantitatively analyzed using Image J software,and the data are presentedMean±SE.Each experiment was performed at least twice with at least duplicate samples.Student'sttest was used to determine the difference between the groups,and aPvalue less than 0.05 was considered to indicate a statistically significant difference.
RESULTS
Screening ESC clones with 7SK truncation
We used a CRISPR/Cas9 system to knockout 7SK from the ESCs,and unexpectedly obtained cells with two novel truncations of 7SK,as shown in Fig.1.The results of sequencing of the PCR product and glycerol gradient analysis confirmed the truncations in the cells.7SK truncation at 128-179 nt,but not at 122-182 nt,caused the release of active P-TEFb from the large complex,which strongly indicates that the nucleotides at 128-179 nt are essential elements for 7SK to form a complex with active P-TEFb.
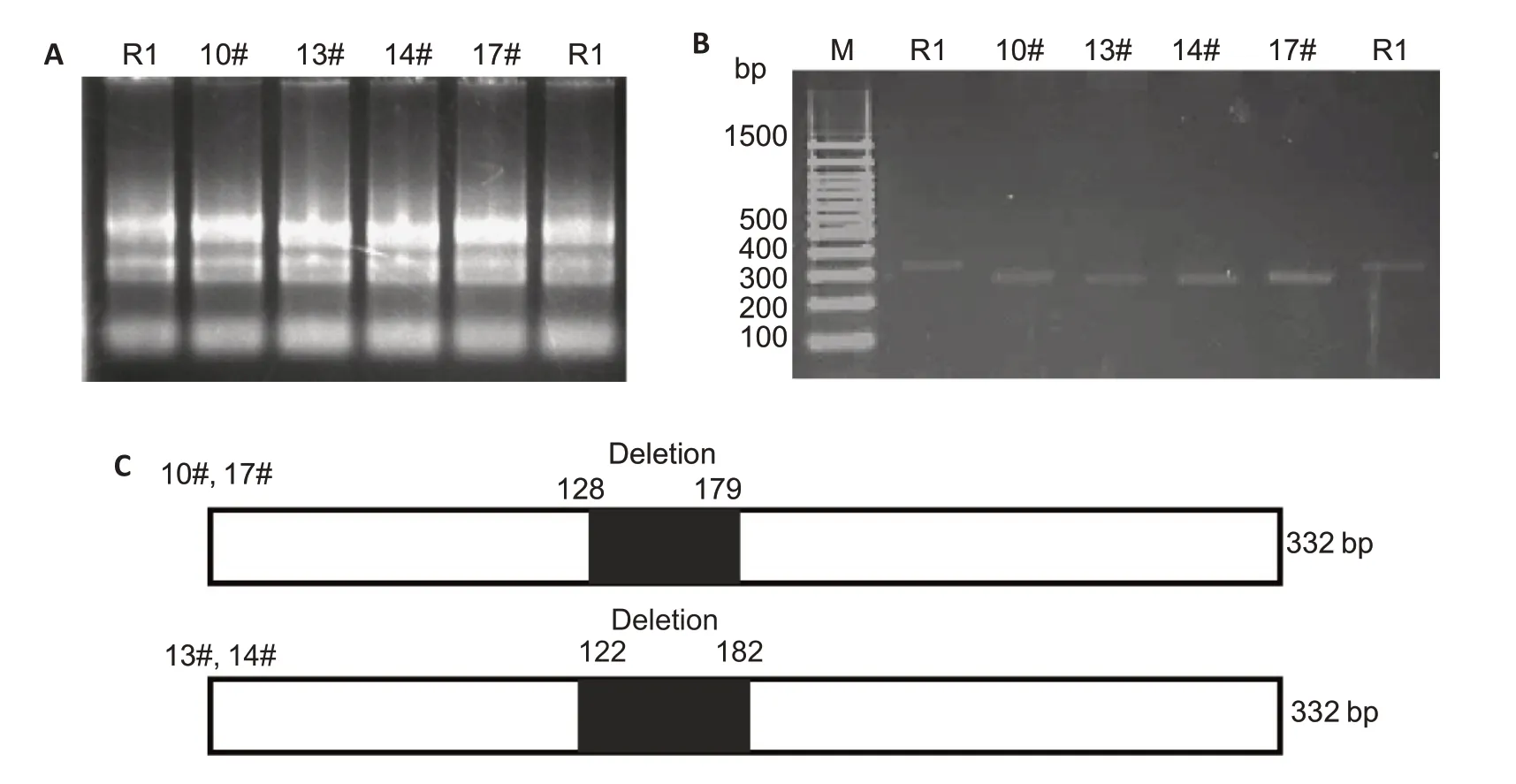
Fig.1 Screening of the positive embryo stem cell (R1) clone with 7SK truncation.A:RNA electropherogram of each single cell clone.B:PCR amplification of cDNA from each single cell clone.C:Schematic presentation of the truncation sites in the single cell clones.
7SK truncation represses proliferation of ESCs
It is generally assumed that an increased expression of active P-TEFb promotes cell growth and proliferation,and thus the observation of global growth failure phenotype in PD patients can be counterintuitive.Compared with the wild-type cells,the ESCs with 7SK truncation exhibited a slower growth rate (P<0.001;Fig.2),as was consistent with the cell phenotype.
7SK truncation downregulates LARP7 and CDC6 in ESCs
We examined the expressions of LARP7 and CDC6 in the ESCs with 7SK truncation using Western blotting.Compared with wild-type cells,the cells with 7SK truncation showed obviously lowered expressions of both LARP7 and CDC6,while the expression of Nanog,the core factor that maintains the stem cell characteristics,did not exhibit significant changes.The expression of minichromosome maintenance 2 (MCM2),which regulates DNA replication,was slightly lowered in the cells with 7SK truncation (Fig.3A).Futhermore,we observed that CDC6 overexpression resulted in significantly accelerated proliferation of R1 cells(Fig.3B,C).As CDC6 is the core component of the pre-replication complex essential for DNA replication,its downregulation by 7SK truncation suggests a high possibility of impaired cell replication mechanism,which is supported by the observation that 7SK truncation caused repression of ESC proliferation.
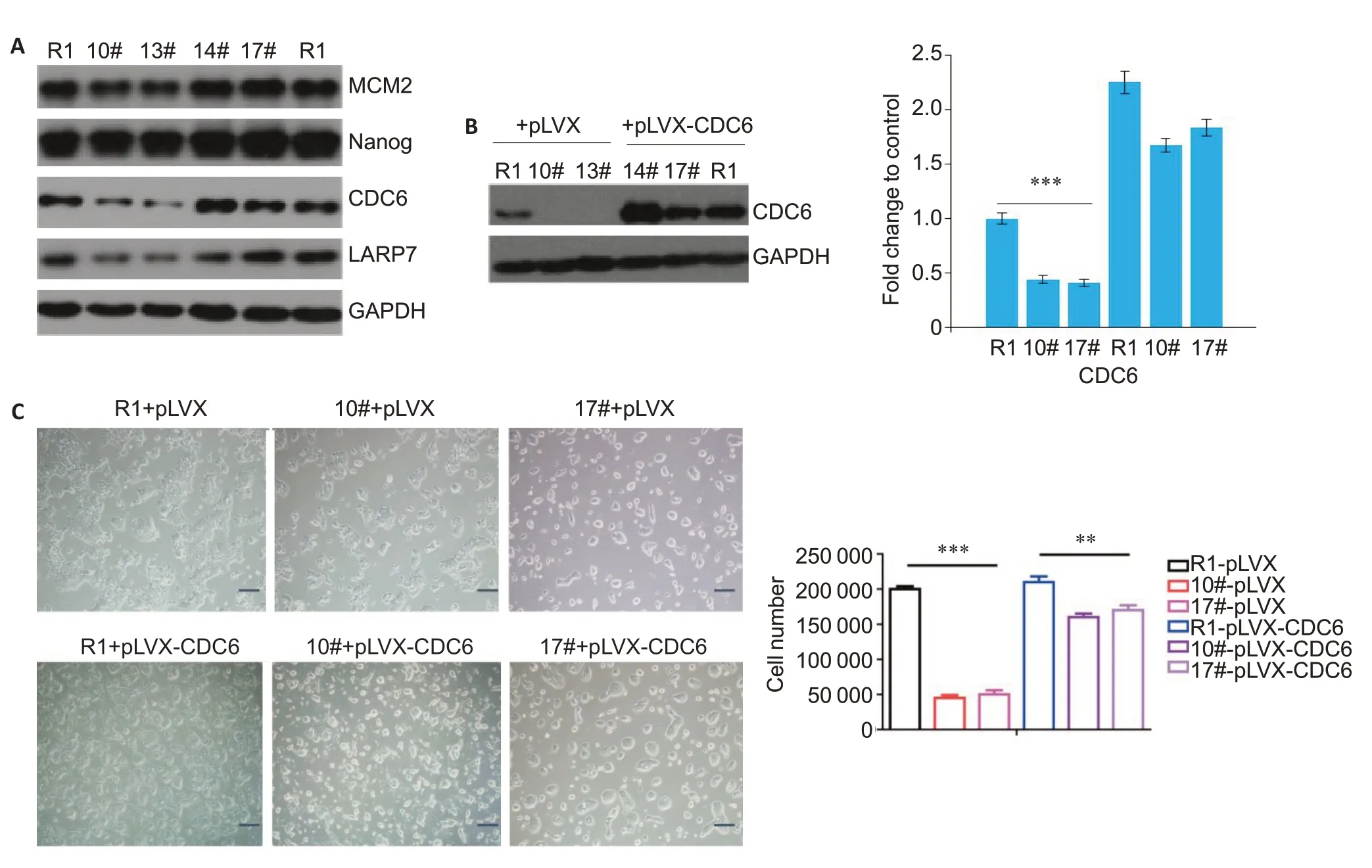
Fig.3 LARP7 and CDC6 expressions are downregulated in ESCs with 7SK truncation.A:Western blotting with indicated antibody for cells with and without 7SK truncation.Representative blots from 3 independent experiments are shown.B:Western blotting for detecting CDC6 expression in cells transfected with pLVX-CDC6 plasmid or blank vector,with GAPDH as the loading control(**P<0.01,***P<0.001 vs control).C:Cell count changes of the ESCs with 7SK truncation transfected with pLVX-CDC6 plasmid or blank vector(Scale bar:100 μm;***P<0.001 vs control).
7SK truncation downregulates CDC6 in ESCs in a CDK9-dependent manner
We found that nucleotide deletion at 128-179 nt in 7SK caused the release of active P-TEFb from the large complex without affecting the activity of CDK9,but increased the level of free CDK9 in the ESCs.Since the cell proliferation process was inhibited,we tested the changes in the core factors of DNA replication:MCM2 and CDC6.MCM2 did not exhibit significant changes,while CDC6 was significantly down-regulated in ESCs with 7SK truncation at 128-179 nt.In order to explore whether CDC6 down-regulation was CDK9-dependent,we knocked down CDK9 with 3 different shRNA constructs in the R1-WT cells,which resulted in obvious downregulation of CDC6 expression (Fig.4A).We also transfected 10# and 17# clones with sh-CDK9-1 construct,and the results of Western blotting showed an obvious down-regulation of CDC6 expression.
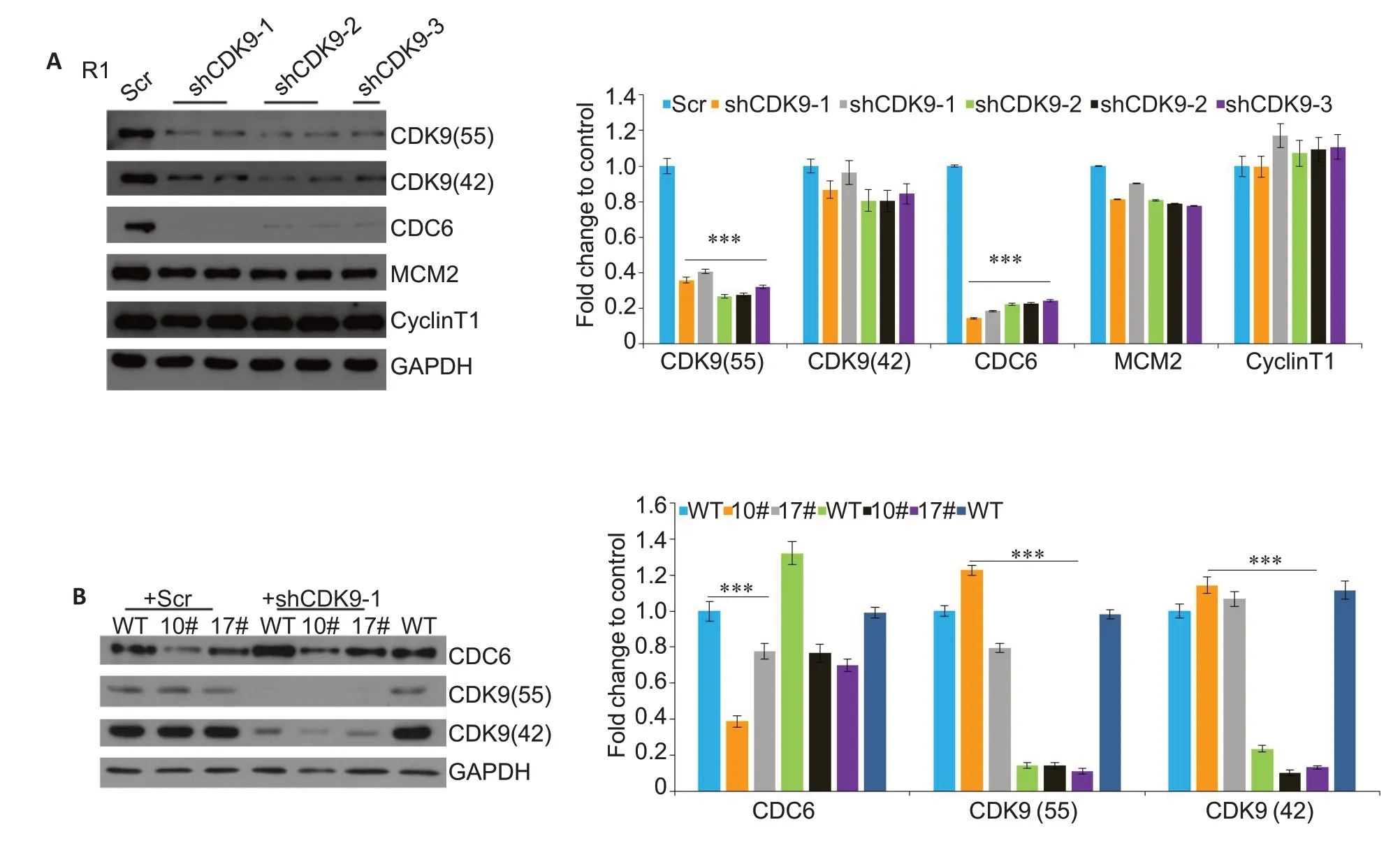
Fig.4 7SK truncation CDK9-dependently downregulates CDC6 in ESCs.A:Knockdown of CDK9 by 3 different shRNA constructs,with a scramble construct (Scr) as the control.The cells were cultured for 3 days after the knockdown before Western blot analyses (***P<0.001 vs control).B:Knockdown of CDK9 by shCDK9-1 constructs in 10#and 17#clones,with the scramble construct(Scr)as the control.The protein levels of CDC6 and CDK9 were determined with Western blotting(***P<0.001 vs control).
DISCUSSION
ESCs offer a unique and excellent system to mimic human diseasesin vitrodue to their abilities to generate various cell types.It has been speculated that defects in PD gene functions may reduce the number of embryonic cells in very early development to result in a smaller size of the embryos in the utero[15].Cultured ESCs can be an ideal system to investigate the underlying mechanisms for this developmental disorder.Accumulating evidence has linked loss-of-function germline mutations of LAPR7 to the pathogenesis of PD[16],but the role of 7SK in PD still remains unclear.A previous study reported that the 3'-terminal is important for 7SK to interact with LARP7[17,18],but our findings show that the nucleotide sequence of 128-179 nt in 7SK is essential for the binding between 7SK and LARP7 and for maintaining the stability of the latter.7SK truncation at 128-179 nt causes the release of active P-TEFb from the complex,as the consequence of a decreased LARP7 protein level.
CDC6 is a core factor during DNA replication,and its loss-of-function mutation is associated with the pathogenesis of PD.LARP7 and HEXIM1,the negative regulatory factors of P-TEFb,were reported to sequester about 70%of P-TEFb in cells[19].Knocking down LARP7 and HEXIM1 can result in the release of active P-TEFb(cyclin T and CDK9)from the complex,but the role of the released active p-TEFb is still unclear[4].We found that the ESCs with CDK9 knockdown showed a reduced expression level of CDC6,whose loss of function has been reported in PD patients[20,22].It is proposed that the defects in PD-related genes are associated with a reduced number of embryonic cells during early embryogenesis,which may account for the global growth failure even in the utero[23,24].We show here that LARP7 deficiency in the ESCs leads to the downregulation of CDC6,which has recently emerged as an important positive regulator of mammalian proliferation[25,26].
Taken together,our results demonstrate that the 128-179 nt sequence in 7SK is essential for LARP7 binding,and the truncation of this sequence causes a decreased expression of CDC6 in ESCs.Our findings shed light on a potential therapeutic target for PD and related developmental diseases.