lncRNA调控畜禽抗病力性状研究进展
杨金艳,刘雪琴,文天琦,孙愉洪,俞英
综 述
lncRNA调控畜禽抗病力性状研究进展
杨金艳1,2,刘雪琴1,2,文天琦1,孙愉洪1,俞英1
1. 中国农业大学动物科学技术学院,北京 100193 2. 云南农业大学动物科学技术学院,昆明 650201
长链非编码RNA (long non-coding RNA, lncRNA)是一类长度大于200个核苷酸的非编码RNA分子。lncRNA虽然不具备蛋白编码能力,但可通过转录调控、转录后调控及表观遗传修饰调控等方式影响基因的表达,进而影响性状的表型。在现代畜牧业生产中,除提高生长发育和产量性状外,研究免疫因子、细胞因子等抗病力相关指标及性状的调控机制,对提高和改善畜禽的健康、福利及公共卫生尤为重要。近年来,利用lncRNA研究鸡()、猪()、牛()等重要畜禽的抗病力性状的调控机制取得了一定进展,为将表观遗传标记应用于动物抗病遗传育种打下了一定的基础。本文介绍了lncRNA的生物学功能和产生机制,着重阐述了lncRNA对畜禽抗病力性状的调控作用及研究进展,以期为lncRNA在畜禽抗病遗传育种方面的研究及应用提供科学依据。
lncRNA;畜禽;抗病力性状
如何提升高产畜禽的抗病力水平,是目前影响畜牧业稳健发展的重要科学问题之一。畜禽抗病力与大部分重要经济性状呈一定程度的负相关。
抗病力主要分广义抗病力和狭义抗病力两类。广义抗病力指抗逆性或抗性,包括机体对疾病的抵抗力,以及对不良气候的耐受力及适应性等[1]。狭义抗病力是指畜禽通过抑制感染,降低病原体的增殖速度[2],不仅涉及机体对某种特殊病原体的抵抗力,也与机体-病原-环境互作密切相关[3]。
畜禽的大部分抗病力性状,如一般性或特殊性抗体水平等属于中低遗传力性状,除遗传因素外,更容易受病原、环境及表观遗传修饰的影响。表观遗传修饰是机体与环境(包括病原体)互作的重要调控机制之一,主要包括DNA甲基化、组蛋白修饰以及非编码RNA (non-coding RNA, ncRNA)等。其中长链非编码RNA (long non-coding RNA, lncRNA)是ncRNA的重要类型之一,可通过染色质水平、转录前和转录后水平调控基因的表达[4~6]。近年来,国内外的研究人员对宿主lncRNA的调控机制及功能关注度日益增加,研究人员开辟了从lncRNA角度研究畜禽疾病发生发展及抗病力水平的新途径,发现lncRNA可以通过参与炎症反应、免疫应答、细胞周期等生物学通路,调控相关基因的表达水平,进而影响畜禽的抗病能力。本文主要综述了lncRNA的生物学功能及作用机制,以及在鸡()、猪()、牛()等主要畜禽抗病力研究领域的新进展和研究策略。
1 lncRNA概述
对lncRNA的研究是一个由表及里,不断深入细化的过程。在20世纪90年代之前,lncRNA被科学界普遍认为是转录的副产物,不具有编码能力,是无用的“垃圾”。20世纪90年代,有研究发现lncRNA能够参与调控表观遗传过程,如H19和XIST等[7,8],lncRNA的功能开始被研究者所关注。2002年,Okazaki等[9]证实lncRNA为转录组的重要组成部分,初步提出了lncRNA的概念。
lncRNA是一类由RNA聚合酶II转录产生、缺乏开放阅读框架且长度大于200 nt的转录物[10]。虽然lncRNA不具有蛋白质编码能力,但分子结构和mRNA相似,具有5′鸟苷帽和3′聚腺苷残基末端,因此又被称为“与mRNA类似的非编码RNA”(mRNA-like non-coding RNA, mlncRNA)[11]。依据lncRNA相对于蛋白质编码基因的位置,可将lncRNA分为5类[12]:正义lncRNA、反义lncRNA、双向lncRNA、内含子lncRNA和基因间lncRNA。
在生物体中,lncRNA存在4种产生模式[12]:(1)蛋白质编码基因突变导致框架结构断裂,从而产生lncRNA (图1A);(2)同一序列复制两次,使相邻的非编码RNA产生重复序列(图1B);(3)转座原件序列插入之后,可产生具有功能的lncRNA (图1C);(4)非编码基因通过逆转录复制,也会产生lncRNA (图1D)。
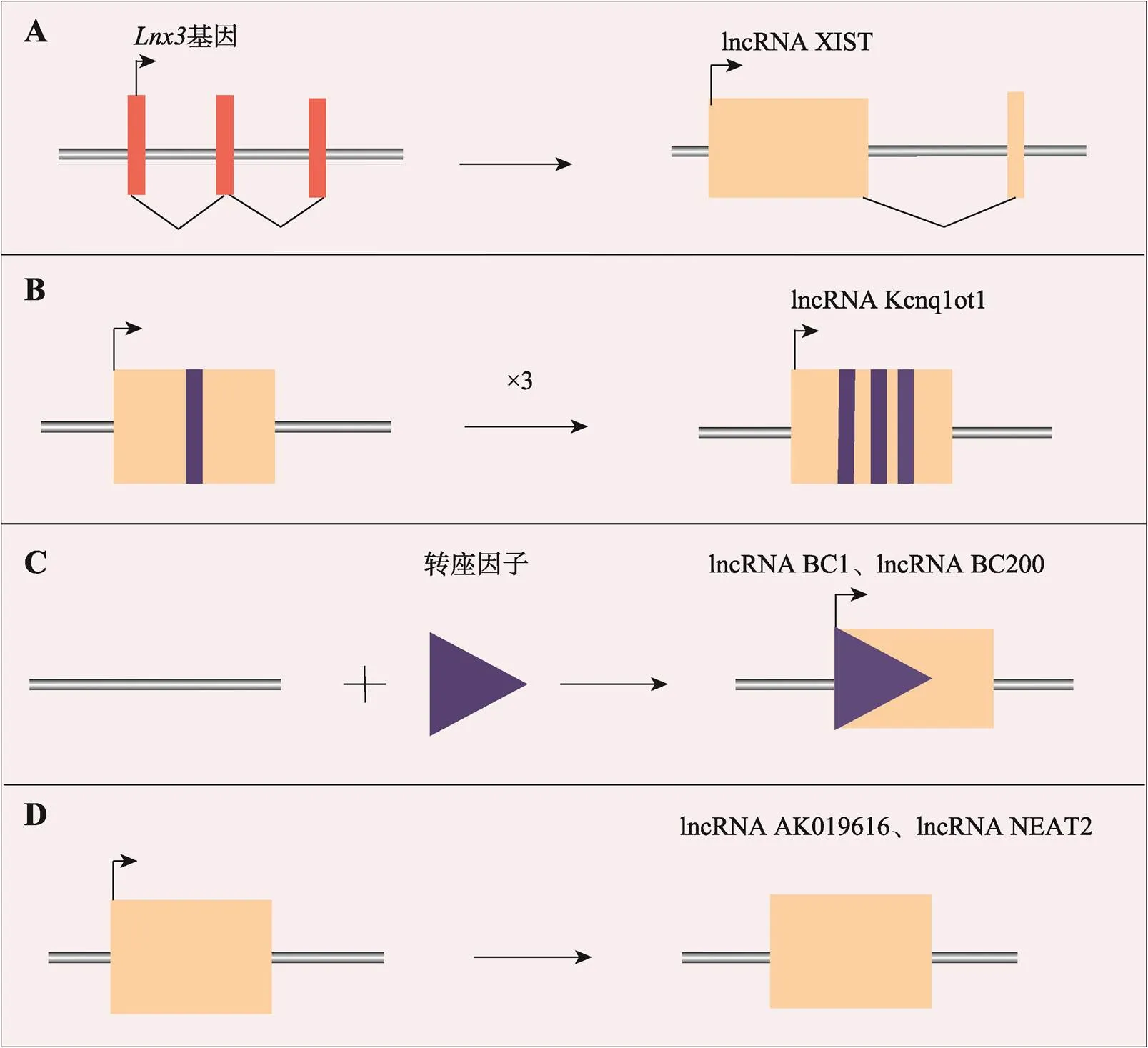
图1 产生lncRNA的主要模式[12]
A:蛋白质编码基因的阅读框发生断裂,产生lncRNA XIST;B:lncRNA Kcnq1ot1的5ʹ区观察到的重复序列;C:lncRNA BC1和 lncRNA BC200来源于转座因子的插入;D:lncRNA AK019616和lncRNA NEAT2由逆转录复制产生。图根据文献[12]修改绘制。
lncRNA可以通过顺式(,临近基因)和反式(,远距离基因)两种方式调控细胞中蛋白编码基因的表达。近年来,lncRNA的研究不断深入,其生物学功能研究从最初的基因组印记、染色质重塑,深入至细胞凋亡周期调控、mRNA的降解、剪接调控和翻译调控等[13~23](表1)。
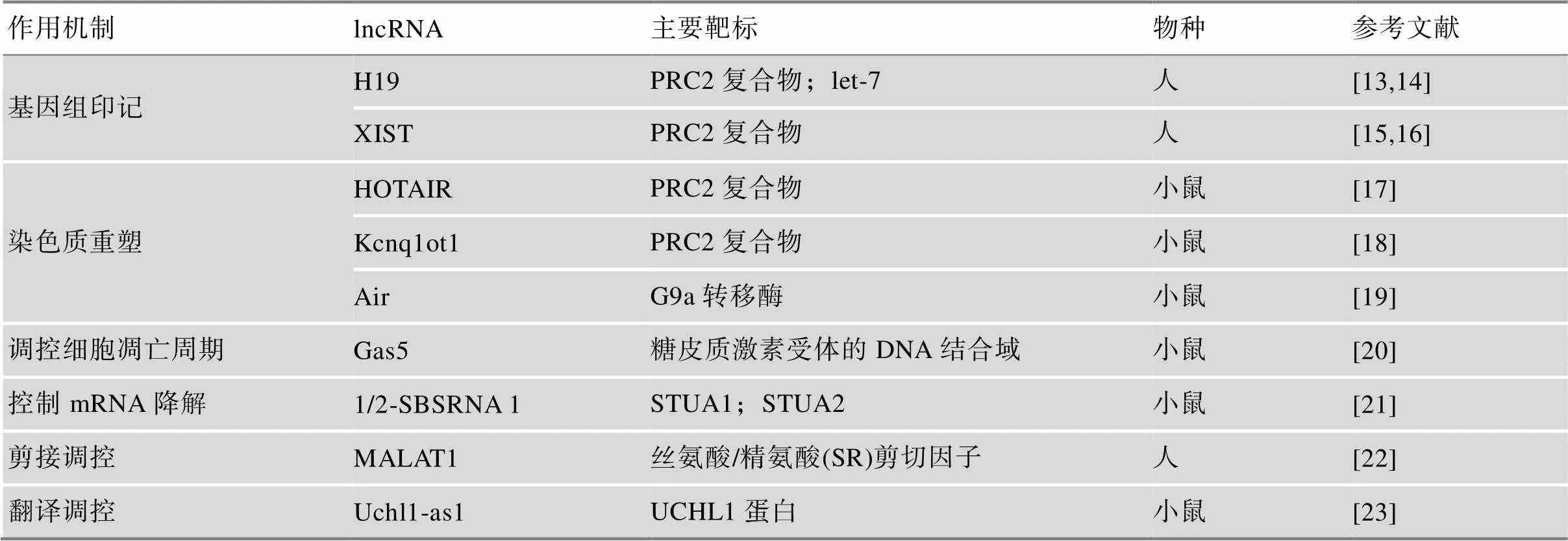
表1 典型lncRNAs的作用机制及关键靶标
2 畜禽抗病力性状相关lncRNAs的筛选和鉴定
在畜禽养殖过程中,动物体若感染细菌或病毒等病原微生物,将引起各类流行性疾病或传染性疾病的发生,导致巨大损失并威胁人类健康。近年来,研究人员探究了病原-lncRNA-宿主相互作用的分子机制,鉴定和挖掘出了一批关键的lncRNAs,为畜禽疾病的诊断、防治和抗病遗传育种提供了重要的分子生物学标记(表2)。
2.1 鸡抗病力性状相关lncRNAs标记
家禽养殖过程中的常见肿瘤性疾病主要包括禽白血病(avian leucosis, AL)和马立克氏病(Marek's disease, MD),分别由禽白血病病毒(avian leukosis virus, ALV)及马立克氏病毒(Marek’s disease virus, MDV)引起。这两种病毒性肿瘤疾病在不同鸡群中的发病率约为15.4%~61%[50,51],双重感染率高达21.92%[52]。J亚型禽白血病病毒(avian leukosis virus subgroup J, ALV-J)感染可引起鸡的肿瘤性疾病,并导致免疫抑制。有研究表明,鸡巨噬细胞(monocyte-derived macrophages, MDMs)感染ALV-J 3小时后,128个lncRNAs和15个miRNAs差异表达;感染36小时后,仅发现30个lncRNAs和8个miRNAs差异表达[24],这说明lncRNA在ALV感染早期比后期更加活跃。感染3小时后的MDMs细胞中,XLOC_672329、ALDBGALG0000001429、XLOC_016500等差异表达的lncRNAs 可以上调免疫相关基因、和的表达水平。与ALV-J未感染组鸡相比,感染组鸡中差异表达lncRNAs的靶基因主要富集于MAP激酶活性、炎症反应等GO条目以及VEGF信号通路、基础转录因子等生物学通路[25]。ALV-J感染组中差异表达的lncRNA TCONS_00060450可以作为潜在的ceRNA,调控关键基因的表达水平。不仅是一种重要的转录调控因子,也可作为肿瘤抑制基因[53],异常表达的会诱发淋巴瘤及恶性肿瘤发生,影响马立克氏肿瘤细胞系MSB1的增殖、迁移和侵袭[54],但及相关lncRNAs的具体调控机制还需要进一步验证。
鸡MD相关报道中,Figueroa等[26]在马立克氏病毒GaHV-2 (也称MDV-1)基因组的TRL/IRL(long terminal repeat/ long internal repeats) 区域,发现一个长7.5 kb的lncRNA——ERL lncRNA (edited repeat- long, long non-coding RNA)。ERL lncRNA在GaHV-2病毒感染和再激活的裂解期和潜伏期均表达,在裂解期被过度编辑(hyperediting),发生A-to-G事件(鸟嘌呤替代腺嘌呤),该过程与干扰素诱导的基因的过表达有关。长基因间非编码RNA (long intergenic non-coding RNAs, lincRNAs)是一种从编码基因之间的DNA序列转录而来的lncRNA,lincRNAs的异常表达与各种类型的癌症和神经系统疾病有关[55]。在鸡中已发现2个lincRNAs (linc- GALMD3和linc-stab1)能够参与调控MD相关的免疫过程[27,28]。linc-GALMD3位于鸡第4条染色体的两个蛋白编码基因之间,可以顺式调控其下游基因的表达[27],同时反式调控MDV感染细胞中其他基因的表达,如、等已有研究证实,下调将会导致鸡后晶状体角膜营养不良[56],诱发鸡虹膜发生病变。因此,linc-GALMD3被认为是关键的调控因子,作为候选的表观遗传标记物,用于MD的预防和诊断。此外,linc-stab1作为另一关键lincRNA,仅在MDV感染潜伏期的MDV抗性鸡品系6中高表达,其表达水平与其邻近的蛋白编码基因的表达水平呈较强的正相关[28],说明linc-satb1可能通过激活基因表达来发挥其抗MDV功能[57]。
2.2 猪抗病力性状相关lncRNA标记
在细菌感染引起的仔猪肠道炎症反应中,产气荚膜梭菌()作为一种食源性人猪共患病病原体[58],可通过产生α肠毒素、β肠毒素来激活免疫和炎症相关的信号通路,增强靶细胞的毒性并诱导超氧化物的产生[59~61]。lncRNA失调将会诱导免疫相关基因表达,进而影响炎性因子和促炎性细胞因子的表达[62],如上调LNC_001066可以显著上调产气荚膜梭菌感染相关基因(、、)的表达水平[30]。同时,这些免疫相关基因的差异表达将会影响入侵过程中仔猪的耐药性和易感性[63]。此外,研究人员还对仔猪感染后,参与免疫应答lncRNA的表达模式和生物学功能进行了深入探索。例如,有研究发现4个lncRNAs (ENSSSCT00000032859、ENSSSCT00000018610、LNC_001066和LNC_001186)在抗性组(resistance groups, IR)及易感组(susceptibility groups, IS)中表达水平存在显著差异[30]。而差异表达lncRNAs的靶向基因在ABC转运蛋白信号转导、MAPK、趋化因子信号和toll样受体等信号通路显著富集,表明这些lncRNAs能够参与调节仔猪感染期间的免疫反应和抗性[31,32,64]。肠毒素大肠杆菌(enterotoxigenic, ETEC)作为另一种致命性肠道病原菌,导致56.2%的仔猪腹泻和24.7%死亡病例[65]。Augustino等[33]全面分析了ETEC感染仔猪小肠上皮细胞的lncRNA和mRNA表达谱,结果显示,LOC102157546和XLOC_025930这两个关键lncRNAs参与 cGMP-PKG信号通路,调控3个黏附表型相关基因(、、)的表达水平,从而影响ETEC-F4ac的黏附表型。
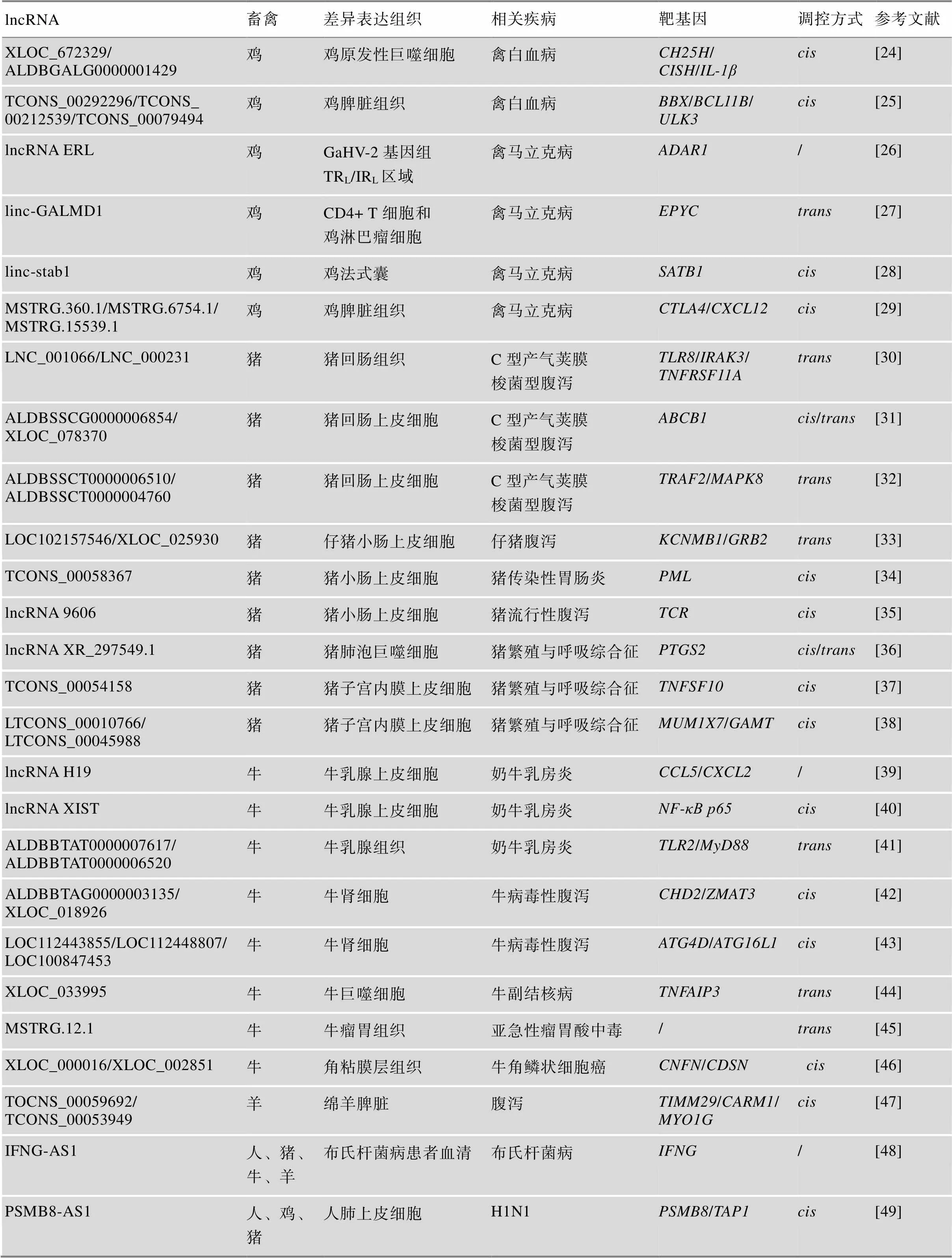
表2 畜禽主要抗病性状相关lncRNAs及其靶基因
在病毒引起的猪炎症反应与免疫反应中,猪繁殖与呼吸综合征是一种由猪繁殖与呼吸综合征病毒(porcine reproductive and respiratory syndrome virus, PRRSV)引起的具有高传染性的急性传染病[66,67]。PRRSV表现出严格的细胞嗜性,主要靶细胞为猪肺泡巨噬细胞(pig alveolar macrophage, PAM)[68]。在PAMs被PPRSV感染的不同时间点,均可发现lncRNA表达谱发生显著变化[69]。PRRSV感染PAMs 9个小时后,环氧合酶-2(COX-2)临近的lncRNAXR_297549.1表达量显著下调,lncRNA XR_297549.1能够顺式和反式调控免疫相关基因的表达水平[36]。另一研究推测[37],PAMs中TCONS_00054158表达上调可能是猪被RNA病毒感染的共同特征,该lncRNA通过上调的表达水平,引发PRRSV感染过程中的细胞凋亡。上述研究为进一步揭示lncRNA调控猪细菌病和病毒病的免疫应答机制提供了理论基础。
2.3 牛抗病力性状相关的lncRNA标记
乳房炎是奶牛最常见的疾病之一,主要由宿主、病原体和环境因素相互作用引起,其中细菌感染是引起奶牛乳房炎的主要原因。大肠杆菌)、金黄色葡萄球菌()和牛分枝杆菌()能够在乳腺组织中快速增殖、黏附并引起炎症,是牛临床和隐性乳房炎的主要传染性病原体[70~72]。同时,宿主的免疫相关信号通路在对抗乳房炎时发挥重要的调控作用,例如NF-κB、MAPK、TLR和JAK-STAT等信号通路[73~76]。以大肠杆菌为主要致病菌的奶牛乳房炎中,脂多糖(lipopolysaccharide, LPS)是主要的毒力因子。LPS通过改变乳腺上皮细胞紧密连接(tight junctions, TJs)的蛋白亚型来破坏血乳屏障[77]。有研究发现,在LPS诱发炎症的组织中,lncRNA H19 (H19)的表达水平显著上调。H19能够促进、、、等炎症因子分泌,同时激活NF-κB通路,促进与β-酪蛋白和紧密连接相关蛋白的表达水平,维持乳腺屏障的完整以防止乳汁成分从乳腺腺泡渗入血清[39]。以金黄色葡萄球菌或大肠杆菌为致病原的乳房炎中,lncRNA XIST (XIST)在MAC-T中表达水平显著上调,XIST通过抑制NF-κB信号通路的激活,阻止炎性细胞因子的产生,并降低NLRP3炎症小体的表达;同时,XIST可以通过负反馈回路来调控NF-κB/NLRP3炎症小体通路,从而介导炎症过程[40]。此外,以牛分枝杆菌为致病原的奶牛乳房炎中,牛分枝杆菌通过和基因激活NF-κB通路,增加IL-1β细胞因子的产生[78]。Ozdemir等[41]研究确定了与和基因显著相关的lncRNAs (ALDBBTAT0000007617和ALDBBTAT0000006520等),这些lncRNAs通过NF-κB和PI3K-Akt通路共同调控牛乳腺组织对牛分枝杆菌感染的免疫应答。
在养殖过程中,若奶牛感染病毒性腹泻病毒(bovine viral diarrhea virus, BVDV),其消化系统会受到严重影响,并出现持续性腹泻和肠炎[79]。BVDV感染牛肾细胞(Madin-darby bovine kidney cells, MDBK)后,随着病毒在细胞中的复制,越来越多的基因被激活并参与免疫应答,同时参与调控的lncRNAs数量也显著增多[42]。BVDV感染过程中,MDBK中差异表达的lncRNAs可以靶向调控等自噬相关基因[42,43]。除了BVDV,奶牛在感染副结核分枝杆菌后,同样也会出现周期性、顽固性腹泻症状[80]。副结核分枝杆菌为牛副结核病的主要病原体,Gupta等[44]发现,由副结核分枝杆菌诱导的牛副结核病中,lncRNA (XLOC_ 033995)能够调控其临近的炎症信号因子的表达水平,并通过参与NF-κB、细胞器裂变等免疫应答相关的通路,影响巨噬细胞对感染的炎症反应进程,最终调控牛副结核病的发病进程。
以上研究结果提示,lncRNA可通过作用于关键靶基因,参与抗病相关基因所在的生物学通路,调控奶牛乳房炎、病毒性腹泻等疾病的发展进程。这些研究结果为奶牛疾病发生的生物标记物挖掘以及奶牛抗病能力的提升提供了新的思路。
2.4 羊抗病力性状相关的lncRNA标记
在羊中高发的羊寄生虫疾病(肝片吸虫病、肺丝虫病、钩虫病)以及腐蹄病等常见疾病与lncRNA关系的研究较少。已有的研究中,Jin等[47]利用大肠杆菌F17菌株饲喂湖羊,并鉴定了对大肠杆菌F17有拮抗或敏感反应个体的lncRNA表达情况,确定了lncRNAs与等6个基因共表达。鉴于的缺失会导致B淋巴细胞硬度降低,进而影响B淋巴细胞的细胞黏附、增殖、吞噬和内吞作用[81],揭示lncRNA对大肠杆菌F17引起的绵羊腹泻具有一定的调控作用。
3 结语与展望
随着分子生物学技术的发展和不同物种转录组数据的积累,lncRNA从最初被认为是转录的“副产物”,到后来被证实能够参与调控人类及动物的多种关键生物学过程。如今越来越多的研究发现,lncRNA可作为猪、鸡、牛等重要畜禽的抗病相关性状的潜在分子标记物,用于疾病的诊断及治疗;同时,lncRNA靶向的基因有望作为畜禽传染性疾病抗性相关的遗传标记,应用于畜禽抗病个体的选择。
然而,对于lncRNA的研究仍然面临一些亟待解决的问题。如,与人类和小鼠等模式动物相比,畜禽的生物数据库中保存的lncRNA转录本数量相对较少且注释信息不完善。因此,未来需进一步完善畜禽基因组lncRNA的注释信息。此外,lncRNA的二级和三级结构的保守性较高,且二级结构中还存在许多未知的“功能性模块”,增加了lncRNA在畜禽抗病育种中的研究难度。可以预见的是,深入探究畜禽抗病相关的lncRNA分子遗传标记物,能够为畜禽的抗病遗传育种提供更加准确的科学数据。
致谢:
感谢中国农业大学动物科学技术学院米思远、唐永杰、刘雪琴、史源钧对本文的修改。
[1] Qian JH, Lian LS. Research progress of breeding for disease resistance in livestock., 2004, 21(1): 53–55.钱锦花, 连林生. 畜禽抗病育种研究进展. 动物科学与动物医学, 2004, 21(1): 53–55.
[2] Best A, White A, Boots M. Maintenance of host variation in tolerance to pathogens and parasites., 2008, 105(52): 20786–20791.
[3] Wang XP, Xu SZ, Gao X, Ren HY, Chen JB. Genetic polymorphism ofgene and correlation with mastitis in cattle., 2007, 34(5): 406–412.
[4] Gong CG, Maquat LE. LncRNAs transactivate STAU1- mediated mRNA decay by duplexing with 3' UTRs via Alu elements., 2011, 470(7333): 284–288.
[5] Yap KL, Li SD, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, Zhou MM. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a., 2010, 38(5): 662–674.
[6] Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, Laurent GS, Kenny PJ, Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of beta-secretase., 2008, 14(7): 723– 730.
[7] Brannan CI, Dees EC, Ingram RS, Tilghman SM. The product of the H19 gene may function as an RNA., 1990, 10(1): 28–36.
[8] Heard E, Mongelard F, Arnaud D, Chureau C, Vourc'h C, Avner P. Human XIST yeast artificial chromosome transgenes show partial X inactivation center function in mouse embryonic stem cells., 1999, 96(12): 6841–6846.
[9] Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, Nikaido I, Osato N, Saito R, Suzuki H, Yamanaka I, Kiyosawa H, Yagi K, Tomaru Y, Hasegawa Y, Nogami A, Schönbach C, Gojobori T, Baldarelli R, Hill DP, Bult C, Hume DA, Quackenbush J, Schriml LM, Kanapin A, Matsuda H, Batalov S, Beisel KW, Blake JA, Bradt D, Brusic V, Chothia C, Corbani LE, Cousins S, Dalla E, Dragani TA, Fletcher CF, Forrest A, Frazer KS, Gaasterland T, Gariboldi M, Gissi C, Godzik A, Gough J, Grimmond S, Gustincich S, Hirokawa N, Jackson IJ, Jarvis ED, Kanai A, Kawaji H, Kawasawa Y, Kedzierski RM, King BL, Konagaya A, Kurochkin IV, Lee Y, Lenhard B, Lyons PA, Maglott DR, Maltais L, Marchionni L, Mckenzie L, Miki H, Nagashima T, Numata K, Okido T, Pavan WJ, Pertea G, Pesole G, Petrovsky N, Pillai R, Pontius JU, Qi D, Ramachandran S, Ravasi T, Reed JC, Reed DJ, Reid J, Ring BZ, Ringwald M, Sandelin A, Schneider C, Semple CAM, Setou M, Shimada K, Sultana R, Takenaka Y, Taylor MS, Teasdale RD, Tomita M, Verardo R, Wagner L, Wahlestedt C, Wang Y, Watanabe Y, Wells C, Wilming LG, Wynshaw-Boris A, Yanagisawa M, Yang I, Yang L, Yuan Z, Zavolan M, Zhu Y, Zimmer A, Carninci P, Hayatsu N, Hirozane-Kishikawa T, Konno H, Nakamura M, Sakazume N, Sato K, Shiraki T, Waki K, Kawai J, Aizawa K, Arakawa T, Fukuda S, Hara A, Hashizume W, Imotani K, Ishii Y, Itoh M, Kagawa I, Miyazaki A, Sakai K, Sasaki D, Shibata K, Shinagawa A, Yasunishi A, Yoshino M, Waterston R, Lander ES, Rogers J, Birney E, Hayashizaki Y, FANTOM Consortium; RIKEN Genome Exploration Research Group Phase I & II Team. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs., 2002, 420(6915): 563–573.
[10] Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: A new frontier of translational research?, 2012, 31(43): 4577–4587.
[11] Kurihara Y, Matsui A, Hanada K, Kawashima M, Ishida J, Morosawa T, Tanaka M, Kaminuma E, Mochizuki Y, Matsushima A, Toyoda T, Shinozaki K, Seki M. Genome- wide suppression of aberrant mRNA-like noncoding RNAs by NMD in Arabidopsis., 2009, 106(7): 2453–2458.
[12] Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs., 2009, 136(4): 629–641.
[13] Schoenherr CJ, Levorse JM, Tilghman SM. CTCF maintains differential methylation at the/locus., 2003, 33(1): 66–69.
[14] Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, Lu LG, Liu CC, Yi JS, Zhang HF, Min W, Bennett AM, Gregory RI, Ding Y, Huang YQ. The imprinted H19 lncRNA antagonizes let-7 microRNAs., 2013, 52(1): 101–112.
[15] Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome., 2008, 322(5902): 750–756.
[16] Wutz A, Rasmussen TP, Jaenisch R. Chromosomal silencing and localization are mediated by different domains of Xist RNA., 2002, 30(2): 167–174.
[17] Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang YL, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIRreprograms chromatin state to promote cancer metastasis., 2010, 464(7291): 1071–1076.
[18] Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C.Kcnq1ot1 antisense noncoding RNA mediates lineage- specific transcriptional silencing through chromatin-level regulation., 2008, 32(2): 232–246.
[19] Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson- Smith AC, Feil R, Fraser P. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin., 2008, 322(5908): 1717–1720.
[20] Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest-and starvation- associated repressor of the glucocorticoid receptor., 2010, 3(107): ra8.
[21] Gong CG, Maquat LE. lncRNAs transactivate STAU1- mediated mRNA decay by duplexing with 3' UTRs via Alu elements., 2011, 470(7333): 284–288.
[22] Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, Blencowe BJ, Prasanth SG, Prasanth KV. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation., 2010, 39(6): 925–938.
[23] Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, Pesce E, Ferrer I, Collavin L, Santoro C, Forrest ARR, Carninci P, Biffo S, Stupka E, Gustincich S. Long non-coding antisense RNA controlstranslation through an embedded SINEB2 repeat., 2012, 491(7424): 454–457.
[24] Dai MM, Feng M, Xie TT, Zhang XQ. Long non-coding RNA and microRNA profiling provides comprehensive insight into non-coding RNA involved host immune responses in ALV-J-infected chicken primary macrophage., 2019, 100: 103414.
[25] Qiu LL, Chang GB, Li ZT, Bi YL, Liu XP, Chen GH. Comprehensive transcriptome analysis reveals competing endogenous RNA networks during avian leukosis virus, subgroup J-induced tumorigenesis in chickens., 2018, 9: 996.
[26] Figueroa T, Boumart I, Coupeau D, Rasschaert D. Hyperediting byof a new herpesvirus lncRNA during the lytic phase of the oncogenic Marek's disease virus., 2016, 97(11): 2973–2988.
[27] Han B, He YH, Zhang L, Ding Y, Lian L, Zhao CF, Song JZ, Yang N. Long intergenic non-coding RNA GALMD3 in chicken Marek's disease., 2017, 7(1): 10294.
[28] He YH, Ding Y, Zhan F, Zhang HM, Han B, Hu GQ, Zhao KJ, Yang N, Yu Y, Mao L, Song JZ. The conservation and signatures of lincRNAs in Marek's disease of chicken., 2015, 5: 15184.
[29] You Z, Zhang QH, Liu CJ, Song JZ, Yang N, Lian L. Integrated analysis of lncRNA and mRNA repertoires in Marek's disease infected spleens identifies genes relevant to resistance., 2019, 20(1): 245.
[30] Huang XY, Sun WY, Yan ZQ, Shi HR, Yang QL, Wang PF, Li SG, Liu LX, Zhao SG, Gun SB. Novel insights reveal anti-microbial gene regulation of piglet intestine immune in response toinfection., 2019, 9(1): 1963.
[31] Huang XY, Sun WY, Yan ZQ, Shi HR, Yang QL, Wang PF, Li SG, Liu LX, Zhao SG, Gun SB. Integrative analyses of long non-coding RNA and mRNA involved in piglet ileum immune response totype C infection., 2019, 9: 130.
[32] Luo RR, Huang XY, Yan ZQ, Gao XL, Wang PF, Yang QL, Wang W, Xie KH, Gun SB. Identification and characterization of MAPK signaling pathway genes and associated lncRNAs in the ileum of piglets infected bytype C., 2020, 2020: 8496872.
[33] Augustino SMA, Xu QL, Liu XQ, Mi SY, Shi LY, Liu YB, Wen H, Wang D, Liu L, Zhang Q, Yu Y. Integrated analysis of lncRNAs and mRNAs reveals key-target genes associated with ETEC-F4ac adhesion phenotype in porcine small intestine epithelial cells., 2020, 21(1): 780.
[34] Ma XL, Zhao XM, Wang KL, Tang XY, Guo JX, Mi M, Qi YP, Chang LL, Huang Y, Tong DW. Identification and analysis of long non-coding RNAs that are involved in inflammatory process in response to transmissible gastroenteritis virus infection., 2019, 20(1): 806.
[35] Chen JN, Zhang CY, Zhang N, Liu GL. Porcine endemic diarrhea virus infection regulates long noncoding RNA expression., 2019, 527: 89–97.
[36] Zeng NF, Wang C, Liu SY, Miao Q, Zhou L, Ge XN, Han J, Guo X, Yang HC. Transcriptome analysis reveals dynamic gene expression profiles in porcine alveolar macrophages in response to the Chinese highly pathogenic porcine reproductive and respiratory syndrome virus., 2018, 2018: 1538127.
[37] Zhang J, Sun P, Gan LP, Bai WJ, Wang ZJ, Li D, Cao YM, Fu YF, Li PH, Bai XW, Ma XQ, Bao HF, Chen YL, Liu ZX, Lu ZJ. Genome-wide analysis of long noncoding RNA profiling in PRRSV-infected PAM cells by RNA sequencing., 2017, 7(1): 4952.
[38] Zhang K, Ge LJ, Dong SS, Liu Y, Wang D, Zhou CY, Ma C, Wang YC, Su F, Jiang YL. Global miRNA, lncRNA, and mRNA transcriptome profiling of endometrial epithelial cells reveals genes related to porcine reproductive failure caused by porcine reproductive and respiratory syndrome virus., 2019, 10: 1221.
[39] Li XZ, Wang H, Zhang YF, Zhang JJ, Qi SP, Zhang Y, Gao MQ. Overexpression of lncRNA H19 changes basic characteristics and affects immune response of bovine mammary epithelial cells., 2019, 7: e6715.
[40] Ma MR, Pei YF, Wang XX, Feng JX, Zhang Y, Gao MQ. LncRNA XIST mediates bovine mammary epithelial cell inflammatory response via NF-κB/NLRP3 inflammasome pathway., 2019, 52(1): e12525.
[41] Özdemir S, Altun S. Genome-wide analysis of mRNAs and lncRNAs ininfected and non-infected bovine mammary gland tissues., 2020, 50: 101512.
[42] Ma QM, Li LY, Tang Y, Fu Q, Liu S, Hu SW, Qiao J, Chen CF, Ni W. Analyses of long non-coding RNAs and mRNA profiling through RNA sequencing of MDBK cells at different stages of bovine viral diarrhea virus infection., 2017, 115: 508–516.
[43] Gao XW, Niu C, Wang Z, Jia S, Han MJ, Ma YY, Guan XT, Wang L, Qiao XY, Xu YG. Comprehensive analysis of lncRNA expression profiles in cytopathic biotype BVDV-infected MDBK cells provides an insight into biological contexts of host-BVDV interactions., 2021, 12(1): 20–34.
[44] Gupta P, Peter S, Jung M, Lewin A, Hemmrich-Stanisak G, Franke A, von Kleist M, Schütte C, Einspanier R, Sharbati S, Bruegge JZ. Analysis of long non-coding RNA and mRNA expression in bovine macrophages brings up novel aspects ofsubspeciesinfections., 2019, 9(1): 1571.
[45] Mahmoudi B, Fayazi J, Roshanfekr H, Sari M, Bakhtiarizadeh MR. Genome-wide identification and characterization of novel long non-coding RNA in Ruminal tissue affected with sub-acute Ruminal acidosis from Holstein cattle., 2020, 44(1): 19–27.
[46] Sabara PH, Jakhesara SJ, Panchal KJ, Joshi CG, Koringa PG. Transcriptomic analysis to affirm the regulatory role of long non-coding RNA in horn cancer of Indian zebu cattle breed Kankrej ()., 2020, 20(1): 75–87.
[47] Jin CY, Bao JJ, Wang Y, Chen WH, Wu TY, Wang LH, Lv XY, Gao W, Wang BZ, Zhu GQ, Dai GJ, Sun W. Changes in long non-coding RNA expression profiles related to the antagonistic effects ofF17 on lamb spleens., 2018, 8(1): 16514.
[48] Gheitasi R, Jourghasemi S, Pakzad I, Sarmadi VH, Samieipour Y, Sekawi Z, Jalilian FA. A potential marker in brucellosis, long non coding RNA IFNG-AS1., 2019, 46(6): 6495–6500.
[49] More S, Zhu ZY, Lin K, Huang CQ, Pushparaj S, Liang YR, Sathiaseelan R, Yang XY, Liu L. Long non-coding RNA PSMB8-AS1 regulates influenza virus replication., 2019, 16(3): 340–353.
[50] Zhang HH, Liu Q, Qiu B, Liu GZ, Cheng ZQ. Mixed infection of ALV-J and MDV in a flock of Shandong free range chickens., 2009, 40(8): 1215–1221.张洪海, 刘青, 邱波, 刘功振, 成子强. 地方柴鸡中J亚群禽白血病与马立克氏病的混合感染. 畜牧兽医学报, 2009, 40(8): 1215–1221.
[51] Qin LT, Gao YL, Pan W, Deng XY, Sun FF, Li K, Qi XL, Gao HL, Liu CN, Wang XM. Investigation of co-infection of ALV-J with REV, MDV, CAV in layer chicken flocks in some regions of China., 2010, 32(2): 90–93.秦立廷, 高玉龙, 潘伟, 邓小芸, 孙芬芬, 李凯, 祁小乐, 高宏雷, 刘超男, 王笑梅. 我国部分地区蛋鸡群ALV-J及与REV、MDV、CAV混合感染检测. 中国预防兽医学报, 2010, 32(2): 90–93.
[52] Wang GJ, Wei P, He XM, Li KR, Xiong LW, Yang L, Mo ML, Tao JH. A survey of the epizootiology of three neoplastic diseases in Guangxi province., 2002, 24(10): 13–15.王桂军, 韦平, 何秀苗, 李康然, 熊丽文, 杨乐, 磨美兰, 陶锦华. 鸡三种肿瘤病在广西的流行病学研究. 中国家禽, 2002, 24(10): 13–15.
[53] Wakabayashi Y, Watanabe H, Inoue J, Takeda N, Sakata J, Mishima Y, Hitomi J, Yamamoto T, Utsuyama M, Niwa O, Aizawa S, Kominami R.is required for differentiation and survival of alphabeta T lymphocytes., 2003, 4(6): 533–539.
[54] Zhao CF, Li X, Han B, You Z, Qu LJ, Liu CJ, Song JZ, Lian L, Yang N. Gga-miR-219b targetingsuppresses proliferation, migration and invasion of Marek's disease tumor cell MSB1., 2017, 7(1): 4247.
[55] Ulitsky I, Bartel DP. LincRNAs: genomics, evolution, and mechanisms., 2013, 154(1): 26–46.
[56] Kim MJ, Frausto RF, Rosenwasser GOD, Bui T, Le DJ, Stone EM, Aldave AJ. Posterior amorphous corneal dystrophy is associated with a deletion of small leucine- rich proteoglycans on chromosome 12., 2014, 9(4): e95037.
[57] Han HJ, Russo J, Kohwi Y, Kohwi-Shigematsu T.reprogrammes gene expression to promote breast tumour growth and metastasis.,2008, 452(7184): 187–193.
[58] Scharff RL. Economic burden from health losses due to foodborne illness in the United States., 2012, 75(1): 123–131.
[59] Waters M, Savoie A, Garmory HS, Bueschel D, Popoff MR, Songer JG, Titball RW, Mcclane BA, Sarker MR. Genotyping and phenotyping of beta2-toxigenicfecal isolates associated with gastrointestinal diseases in piglets., 2003, 41(8): 3584– 3591.
[60] Sayeed S, Uzal FA, Fisher DJ, Saputo J, Vidal JE, Chen Y, Gupta P, Rood JI, Mcclane BA. Beta toxin is essential for the intestinal virulence oftype C disease isolate CN3685 in a rabbit ileal loop model., 2008, 67(1): 15–30.
[61] Duan X, Nauwynck HJ, Favoreel HW, Pensaert MB. Identification of a putative receptor for porcine reproductive and respiratory syndrome virus on porcine alveolar macrophages., 1998, 72(5): 4520–4523.
[62] Carpenter S, Aiello D, Atianand MK, Ricci EP, Gandhi P, Hall LL, Byron M, Monks B, Henry-Bezy M, Lawrence JB, O'Neill LAJ, Moore MJ, Caffrey DR, Fitzgerald KA. A long noncoding RNA mediates both activation and repression of immune response genes., 2013, 341(6147): 789–792.
[63] Laine AL, Burdon JJ, Nemri A, Thrall PH. Host ecotype generates evolutionary and epidemiological divergence across a pathogen metapopulation., 2014, 281(1787): 20140522.
[64] Yan ZQ, Huang XY, Sun WY, Yang QL, Shi HR, Jiang TT, Li SG, Wang PF, Gun SB. Analyses of long non-coding RNA and mRNA profiling in the spleen of diarrheic piglets caused bytype C., 2018, 6: e5997.
[65] Li YH, Qiu XT, Li HJ, Zhang Q. Adhesive patterns ofF4 in piglets of three breeds., 2007, 34(7): 591–599.
[66] Lunney JK, Fang Y, Ladinig A, Chen NH, Li YH, Rowland B, Renukaradhya GJ. Porcine reproductive and respiratory syndrome virus (PRRSV): Pathogenesis and interaction with the immune system., 2016, 4: 129–154.
[67] Li YF, Wang XL, Bo KT, Wang XW, Tang B, Yang BS, Jiang WM, Jiang P. Emergence of a highly pathogenic porcine reproductive and respiratory syndrome virus in the Mid-Eastern region of China., 2007, 174(3): 577– 584.
[68] Duan X, Nauwynck HJ, Pensaert MB. Effects of origin and state of differentiation and activation of monocytes/ macrophages on their susceptibility to porcine reproductive and respiratory syndrome virus (PRRSV)., 1997, 142(12): 2483–2497.
[69] Badaoui B, Rutigliano T, Anselmo A, Vanhee M, Nauwynck H, Giuffra E, Botti S. RNA-sequence analysis of primary alveolar macrophages afterinfection with porcine reproductive and respiratory syndrome virus strains of differing virulence., 2014, 9(3): e91918.
[70] Bar-Gal GK, Blum SE, Hadas L, Ehricht R, Monecke S, Leitner G. Host-specificity ofcausing intramammary infections in dairy animals assessed by genotyping and virulence genes., 2015, 176(1–2): 143–154.
[71] Bayoumi FA, Farver TB, Bushnell B, Oliveria M. Enzootic mycoplasmal mastitis in a large dairy during an eight-year period., 1988, 192(7): 905–909.
[72] Fox LK, Gay JM. Contagious mastitis., 1993, 9(3): 475–487.
[73] He XX, Liu WJ, Shi MY, Yang ZT, Zhang XC, Gong PT. Docosahexaenoic acid attenuates LPS-stimulated inflammatory response by regulating the PPARγ/NF-κB pathways in primary bovine mammary epithelial cells., 2017, 112: 7–12.
[74] Wang JJ, Guo CM, Wei ZK, He XX, Kou JH, Zhou ES, Yang ZT, Fu YH. Morin suppresses inflammatory cytokine expression by downregulation of nuclear factor-κB and mitogen-activated protein kinase (MAPK) signaling pathways in lipopolysaccharide-stimulated primary bovine mammary epithelial cells., 2016, 99(4): 3016–3022.
[75] De Schepper S, De Ketelaere A, Bannerman DD, Paape MJ, Peelman L, Burvenich C. The toll-like receptor-4 (TLR-4) pathway and its possible role in the pathogenesis ofmastitis in dairy cattle., 2008, 39(1): 5.
[76] Usman T, Yu Y, Liu C, Wang X, Zhang Q, Wang YC. Genetic effects of single nucleotide polymorphisms inandgenes on susceptibility of Chinese Holsteins to mastitis., 2014, 41(12): 8293– 8301.
[77] Kobayashi K, Oyama S, Numata A, Rahman MM, Kumura H. Lipopolysaccharide disrupts the milk-blood barrier by modulating claudins in mammary alveolar tight junctions., 2013, 8(4): e62187.
[78] Wang Y, Liu SL, Li Y, Wang Q, Shao JR, Chen Y, Xin JQ.-derived lipid-associated membrane proteins activate IL-1β production through the NF-κB pathway viaand., 2016, 55: 111–118.
[79] Perdrizet JA, Rebhun WC, Dubovi EJ, Donis RO. Bovine virus diarrhea—clinical syndromes in dairy herds., 1987, 77(1): 46–74.
[80] Sweeney RW. Pathogenesis of paratuberculosis., 2011, 27(3): 537–546.
[81] López-Ortega O, Ovalle-García E, Ortega-Blake I, Antillón A, Chávez-Munguía B, Patiño-López G, Fragoso-Soriano R, Santos-Argumedo L. Myo1g is an active player in maintaining cell stiffness in B-lymphocytes., 2016, 73(5): 258–268.
Progress on lncRNA regulated disease resistance traits in domesticated animals
Jinyan Yang1,2, Xueqin Liu1,2, Tianqi Wen1, Yuhong Sun1, Ying Yu1
Long non-coding RNA (lncRNA) is a class of non-coding RNAs with a length greater than 200 nucleotides. Although lncRNAs do not have any protein coding capability, they can affect the phenotypes of traits by influencing gene expression through transcriptional regulation, post-transcriptional regulation, and epigenetic modification. In modern animal husbandry production, besides increasing growth and yield traits, investigations on the regulation mechanisms of immune factors, cytokines and other disease resistance-related indicators and traits are particularly important for improving the health and welfare of domesticated animals as well as public health. In recent years, researchers have made significant progress in understanding the regulatory mechanisms of lncRNA on the disease resistance traits of chickens (), pigs (), cattle (and other important domesticated animals, thereby laying the basic foundation for the translational application of epigenetic markers in breeding of animals with disease resistance. In this review, we briefly introduce the biological functions and the origins of lncRNAs, then focus on the research progress on the regulatory effects of lncRNAs on disease resistance traits of domesticated animals, and thus providing the scientific basis for the research of lncRNA and its application in the breeding of disease-resistant animals.
lncRNA; domesticated animals; disease-resistance traits
2021-01-10;
2021-06-16
国家自然科学基金项目(编号:31961143009,31272420),北京市自然科学基金项目(编号:6182021),北京市奶牛产业创新团队项目(编号:BAIC06)和国家奶牛产业技术体系项目(编号:CARS-36)资助[Supported by the National Natural Science Foundation of China (Nos.31961143009, 31272420), the Beijing Natural Science Foundation (No. 6182021), the Beijing Dairy Industry Innovation Team (No. BAIC06) and the National Dairy Industry Technology System Project (No. CARS-36)]
杨金艳,本科生,专业方向:动物科学。E-mail: cauyangjinyan@163.com
刘雪琴,在读博士研究生,研究方向:动物分子数量遗传学。E-mail: cauliuxueqin@163.com
杨金艳和刘雪琴并列第一作者。
俞英,博士,教授,博士生导师,研究方向:动物抗病遗传育种及表观遗传调控机理。E-mail: yuying@cau.edu.cn
10.16288/j.yczz.20-230
2021/6/28 13:58:51
URI: https://kns.cnki.net/kcms/detail/11.1913.R.20210628.1047.002.html
(责任编委: 李明洲)

