Wild-type MIC Distribution and Epidemiological Cut-off Value and Resistant Characteristics of Colistin Against Escherichia Coli from Chickens
Liu Yu-hao, Hu Wan-jun, Tian Er-jie, Muhammad Ishfaq, Zhang Xiu-ying, , Chen Chun-li, , and Li Ji-chang, *
1 College of Veterinary Medicine, Northeast Agricultural University, Harbin 150030, China
2 Yichun Customs District P. R. China, Yichun 153000, Heilongjiang, China
3 Heilongjiang Key Laboratory for Animal Disease Control and Pharmaceutical Development, Harbin 150030, China
Abstract: The aim of the present study was to investigate minimum inhibitory concentration (MIC) distributions by broth microdilution (BMD) method and to determine the preliminary epidemiological cut-off value (ECV) of colistin by epidemiological cut-off (ECOFF) finder against E. coli from chickens in China. Anal swabs were collected from chicken farms in China. BMD method was used to measure MIC50 and MIC90 of colistin which were 2 and 4 µg • mL-1, respectively. MIC frequency distributions for colistin were used to estimate preliminary ECV (8 µg • mL-1). High percentages of resistance to ampicillin (94.12%), nalidixic acid (94.12%), enrofloxacin (94.12%), tetracycline (94.12%), ciprofloxacin (88.24%), florfenicol (88.24%), neomycin (64.71%),gentamicin (58.82%), levofloxacin (58.82%), doxycycline (88.24%) and cefalexin (76.47%) were found. In addition, low percentages of resistance to amikacin (5.88%), spectinomycin (17.65%) and fosfomycin (41.18%) were noted. Notably, amoxicillin, sulfisoxazole and trimethoprim resulted in a 100 % resistance generation efficacy rate. Prevalence of mcr-1 in E. coli (9/17) in chromosomal DNA was higher than mcr-4 (2/17) gene, and mcr-1 (5/17) was higher than mcr-4 (3/17) in plasmid.
Key words: E. coli, broth microdilution (BMD) method, epidemiological cut-off value (ECV), mcr-1, mcr-4, colistin
Introduction
Colistin (also known as polymyxin E) is a polypeptide antibiotic that is originally isolated in 1947 from the soil bacteriumPaenibacillus polymyxasubsp.Colistinus. Colistin and polymyxin B belong to the class of polymyxin, which is one of the primary classes of antibiotics with activity against most gram-negative bacteria. Colistin has historically played a minor role as an anti-infective therapy due to its nephrotoxicity,as well as the availability of alternative antimicrobial agents (Falagaset al., 2009). However, the emergence of multidrug-resistant (MDR) Enterobacteriaceae and the lacking of new antibiotics in the market bring serious consequences to the use of antibiotics. Colistin is considered as the last-line therapy to treat against multi-drug resistant gram negative bacterial (MDRGNB) infections (Liet al., 2006; Olaitanet al., 2014).
In recent years, multiple studies have indicated that the prevalence of colistin resistance has increased rapidly among Enterobacteriaceae (Poirelet al., 2017).So, it is necessary to detect the sensitivity ofE. colito colistin and establish preliminary epidemiological cut-off value (ECV) for the rational use of colistin.The authoritative organizations to set the breakpoint include the Clinical and Laboratory Standards Institute (CLSI) in the US and European Committee on Resistant Characteristics Testing (EUCAST)in the Europe. In 2017, a CLSI and EUCAST Join Working Group recommended broth microdilution(BMD) method, as the reference method for testing colistin (CLSI, 2015). EUCAST advised against the use of gradient and disc diffusion tests for colistin.In order to facilitate the drug resistance monitoring and provide scientific basis for the risk assessment of drug resistance, it is necessary to detect the sensitivity of colistin againstE. coliand establish preliminary ECV, recently. The resulting minimum inhibitory concentration (MIC) frequency distributions for antimicrobials are used to estimate preliminary ECV which separates isolates with typical wildtype susceptibility from isolates with decreased nonwild-type susceptibility (Turnidgeet al., 2006; Schonet al., 2009) by epidemiological cut-off (ECOFF)finder software (Turnidgeet al., 2007), which is based on the principle of non-linear regression to simplify the process of wild-type critical value formulation by EUCAST (Leclercqet al., 2013). At present, ECOFF finder software is gradually applied in various studies.EUCAST publishes MIC distribution data and ECOFF value of different bacterial species or different drugs on its official website (http://www.eucast.org/), and ECOFF value of a drug can be found for the corresponding bacteria as needed.
The wide use of colistin in veterinary medicine for the control of MDR infection and for prophylaxis purposes causes a significant increase in colistin resistance. MDRE. coliposes a great challenge for public health in recent decades. Multiple resistance of colistin againstE. colifrom chickens is tested by Kirby-Bauer (K-B) method. In order to investigate the possible contribution of plasmid encodedmcr-1 andmcr-4 genes in the resistance development, colistin resistant genesmcr-1 andmcr-4 are examined by PCR.Until 2015, all the characterized colistin resistance mechanisms were chromosomally encoded, and thus only limited vertical transmission of resistance was envisioned (Olaitanet al., 2014). However, the discovery of the plasmid-borne phosphoethanol-amine transferase resistance determinantmcr-1 reveals a mechanism for horizontal spread (Liuet al., 2016).Several othermcrhomologs (mcr-2,mcr-3,mcr-4,mcr-5,mcr-6,mcr-7 andmcr-8) are subsequently identified in Enterobacteriaceae until now (Garcia-Graellset al., 2018). The genemcr-4 is firstly characterized inSalmonellaandE. colifrom European countries and further detected inEnterobacter cloacaeandE. coliisolates from Asia (Singapore and Japan)(Jeanettet al., 2018). ECV for colistin has not been described forE. coliin China. The purpose of this study was to investigate MIC distributions by BMD and determine the preliminary ECV of colistin by ECOFF finder againstE. colifrom chickens in China. Besides, multiple resistance of colistin againstE. colifrom chickens was tested by K-B method.Additionally, colistin resistant genesmcr-1 andmcr-4 were examined by PCR.
Materials and Methods
Sample collection and identification
Sterile swabs were used to collect anal swabs from large scale farms (chicken) in 10 provinces of China.To isolateE. coli, anal samples were suspended in Mueller Hinton (MH) broth and enriched overnight at 37℃, then a loopful of the turbid solution from overnight nutrient broth culture was plated onto eosin methylene blue (EMB) agar and MacConkey agar plates. Each plate was incubated at 37℃ for 20-24 h and the positive colonies were selected. Suspected colonies ofE. coliwere aseptically sub cultured on to freshly prepared EMB and MacConkey agar plates for pure strains, and then were incubated at 37℃ for 20-24 h.E. coliwas identified using standard microbiology identification technique which using PCR ampli-fication of 16S rRNA gene using the commonly used primer pair 27F/1492R (Williaomet al., 1991).Besides, the determinedE. coliwas randomly selected for biochemical identification.
MIC assays by BMD
MICs of colistin were determined by the ISO-20776(CLSI, 2010) standard (BMD) method using colistin over a range of dilutions from 0.25 µg • mL-1to 128 µg • mL-1. Briefly, 100 µL of antibiotic suspensions were added at increasing two-fold concentrations as the followings: 0.25, 0.5, 1, 2, 4, 8, 16, 32, 64 and 128 µg • mL-1for colistin. Then, eachE. colistrain was re-suspended from plates in MH broth to a concentration of 0.5 McFarland standard, was diluted 1 000-fold and then distributed into 96-well plates at a concentration of 100 µL • well-1. MICs were determined as the lowest antibiotic concentrations for which no visible growth ofE. coliwas detected after 16-18 h at 37℃. Wells containing 200 µL of MH broth were used as negative and 200 µL of bacterial suspension without any antibiotic was the positive growth controls.E. coliATCC 25922 strain was used as the quality control strain.
Determination of preliminary ECV
Skewness and kurtosis were 0.381 and 0.565, respectively. MIC distribution could be considered as close to normal distribution.
CLSI recommended a sample size of N>300 isolates for ECV determination. Histograms of the MIC distribution by BMD method were generated and ECV was estimated by iterative non-linear regression on expanding subsets using ECOFF finder tool and additional visual inspection. ECOFF finder software was used to determine ECV. ECOFF finder calculated four ECVs that included 95.0%, 97.5%, 99.0% and 99.9% of the isolates in the susceptible population.An ECV of 95% was deemed susceptible (S) and an ECV between 95%-99.9% was deemed intermediate (I)(Ismailet al., 2018). Optimum fits were found when the difference between the estimated and true number of isolates in the fitted subset was minimal. ECV was the highest MIC that defined the wild type population,those isolates with no decreased susceptibility for the antimicrobial agent were evaluated. MIC of 95%confidence interval was selected as ECV.
Bacterial susceptibility testing
Six colistin-resistantE. coli(colistin MIC>128 µg • mL-1)were screened and 11E. colistrains (colistin MIC=8 µg • mL-1) were used by K-B method. Briefly, the antibiotic disks were aseptically placed at a distance of 15 mm apart on MH agar plates that were already swabbed or inoculated withE. coliisolates (adjusted to 0.5 McFarland turbidity standards). Each plate was incubated at 37℃ for 18 h.E. coliATCC 25922 strain was used as the quality control strain for the multiple resistance test. Inhibition zones were interpreted according to CLSI criteria. The antimicrobials used were ampicillin, nalidixic acid, enrofloxacin, tetracycline, ciprofloxacin, florfenicol, doxycycline, amikacin, spectinomycin, gentamicin, neomycin, levofloxacin, fosfomycin, amoxicillin, sulfisoxazole,trimethoprim and cefalexin and were commercially obtained from Hangzhou Microbial Reagent Co., Ltd(China).
PCR amplification of mcr-1 and mcr-4 genes
The presence of plasmid and chromosome DNA encoded resistance genes,mcr-1 andmcr-4 genes were examined by PCR for screened 17E. coli.mcr-1 was confirmed by PCR using the primers formcr-1-forward 5'-CGGTCAGTCCGTTTGTTC-3'; reverse 5'-CTTGGTCGGTCTGTAGGG-3'-amplicon size 309 bp as previously described by Liuet al(2016).mcr-4 used the primers formcr-4-reverse 5'-CT GCTGACTGGGCTATTACCGTCAT-3'; forward AATTGTCGTGGGAAAAGCCGC-amplicon size 1 062 bp (Teoet al., 2018). Amplification included initial denaturation at 94℃ for 3 min, followed by 30 cycles of DNA denaturation at 94℃ for 30 s,annealing at 52℃ for 30 s, and extension at 72℃ for 1 min.
Results
E. coli isolations and identifications
A total of 1 250E. coliwere collected and identified,which included 199 (15.92%), 53 (4.24%), 402(32.16%), 98 (7.84%), 17 (1.36%), 2 (0.16%), 114(9.12%), 204 (16.32%), 85 (6.80%) and 76 (6.08 %)strains ofE. coliisolates from Heilongjiang, Jilin,Liaoning, Henan, Yunnan, Shandong, Sichuan, Hubei provinces, Inner Mongolia Autonomous Region and Shanghai City during 2016-2018 in Table 1.
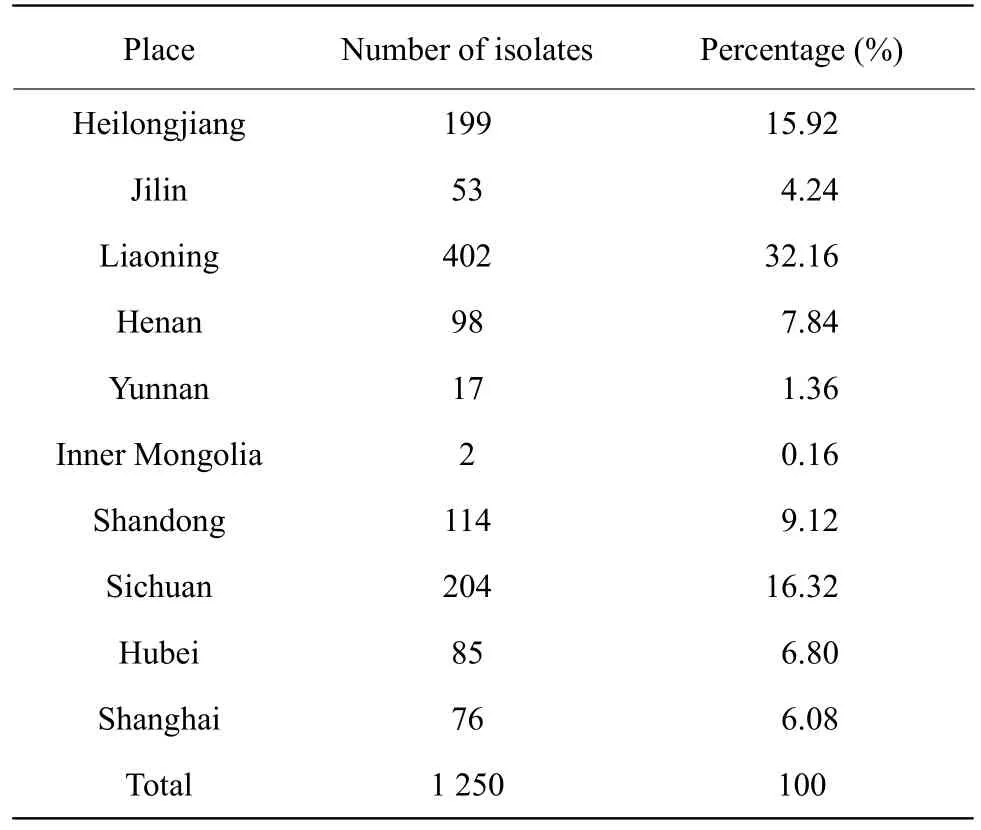
Table 1 Sampling places of E. coli
MIC Distribution
MICs of colistin against 1 250E. coliwere tested by BMD. The range of colistin MICs, according to the BMD method, for all the 1 250 isolates, is presented in Fig. 1 and Table 2. The distribution of each MIC(0.25, 0.5, 1, 2, 4, 8, 16, 32, 64 and 128 µg • mL-1)among tested isolates was as the followings: 0.25µg • mL-1(0.80%), 0.5 µg • mL-1(6.64%), 1 µg • mL-1(25.60%), 2 µg • mL-1(52.40%), 4 µg • mL-1(13.04%),8 µg • mL-1(1.04 %), 16 µg • mL-1(0%), 32 µg • mL-1(0%), 64 µg • mL-1(0%), 128 µg • mL-1(0%) and MIC>128 µg • mL-1(0.48%). MIC50 and MIC90 were 2 and 4 µg • mL-1, respectively.
Determination of preliminary ECV
Both skewness and kurtosis value were 0.381 and 0.565 less than 1. MICs of colistin were brought into ECOFF finder software, and MIC results were converted to log2 MIC as shown in Table 2. Nonlinear regression fitting and the fitting distribution are shown in Fig. 2.The difference between the fitting value and the true value was shown as "ABS (N-Nest)" in Table 3.As shown in Fig. 2, when MIC was no more than 8 µg • mL-1displacement, the difference between the true value and the fitting value was 31. At this point,the difference was the smallest. The upper limit of the distribution of wild type strains within different confidence intervals of 95%, 97.5%, 99.0%, 99.5%and 99.9% was obtained. The fitting results are shown in Table 3. MIC of 95% of the confidence interval was selected as the wild-type critical value. The preliminary ECV of colistin againstE. coliwas 8 µg • mL-1in this study.

Fig. 1 Histogram of colistin BMD MIC distribution (µg • mL-1), N=1 250
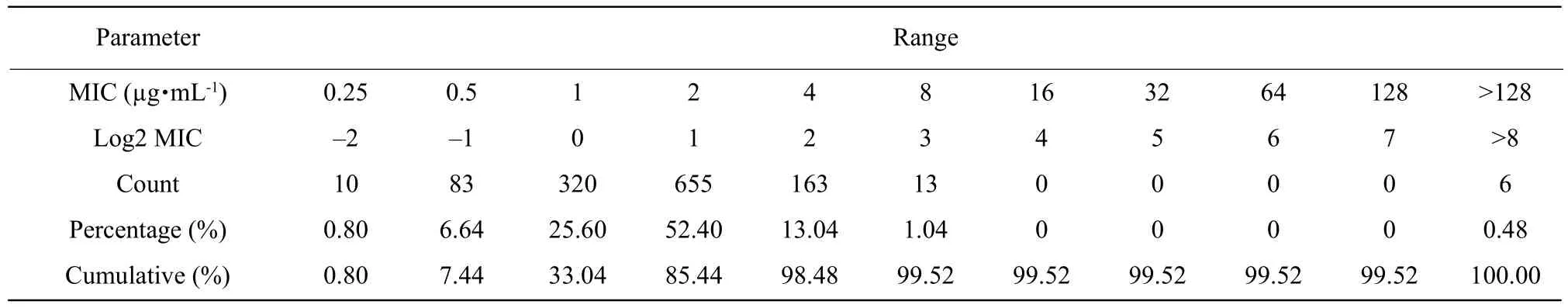
Table 2 MICs of colistin against 1 250 strains of E. coli

Fig. 2 ECV estimation using iterative non-linear regression on expanding (ECOFF finder software) for colistin on BMD, N=1 244
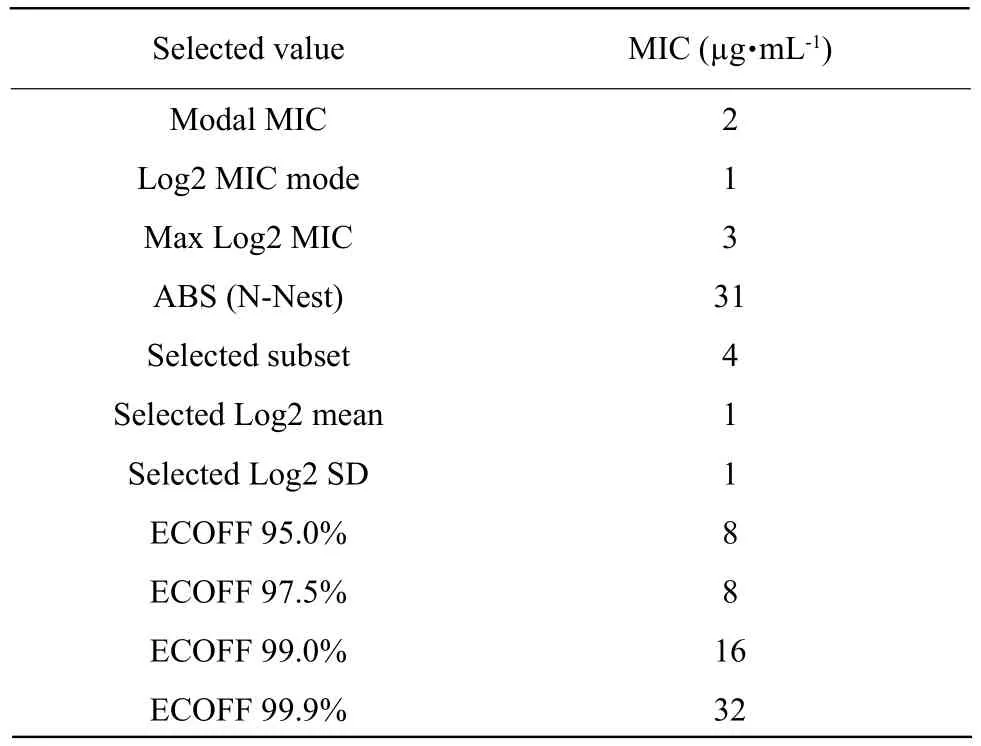
Table 3 Results of ECOFF finder simulation for colistin against E. coli
Susceptibility testing result
A high rate of resistance was noted among the testE. coliisolates as shown in Table 4. It had been noted thatE. coliisolates were highly resistant to amoxicillin, sulfisoxazole and trimethoprim. Overall, the percentage resistance ofE. coliisolates to the tested antibiotics showed that the organism was resistant to ampicillin (94.12%), nalidixic acid (94.12%), enrofloxacin (94.12%), tetracycline (94.12%), ciprofloxacin (88.24%), florfenicol (88.24%), doxycycline(88.24%), cefalexin (76.47%), gentamicin (58.82%),neomycin (64.71%) and levofloxacin (58.82%).However, amikacin, spectinomycin and fosfomycin also showed more susceptibility to the tested isolates at a rate of 5.88%, 17.65% and 41.18%, respectively.The results showed that low susceptibilityE. colistrains of colistin were also resistant to most antibiotics.
Results of positive mcr-1 and mcr-4 genes
In chromosome DNA,mcr-1 was detected in No.9(52.9%) ofE. coliselected, of which No.3 and No.6 were from MIC>128 µg • mL-1(6) and MIC=8 µg • mL-1(11), respectively. In plasmid,mcr-1 was detected in No.5 (29.4 %), of which No.1 and No.4 were from MIC>128 µg • mL-1(6) and MIC=8 µg • mL-1(11),respectively. However,mcr-4 was positive only in colistin resistantE. coliwith MIC>128 µg • mL-1. The genemcr-4 was detected in No.5 ofE. coliscreened,of which No.3 and No.2 were from plasmid and chromosome DNA, as shown in Table 5 and Fig. 3.
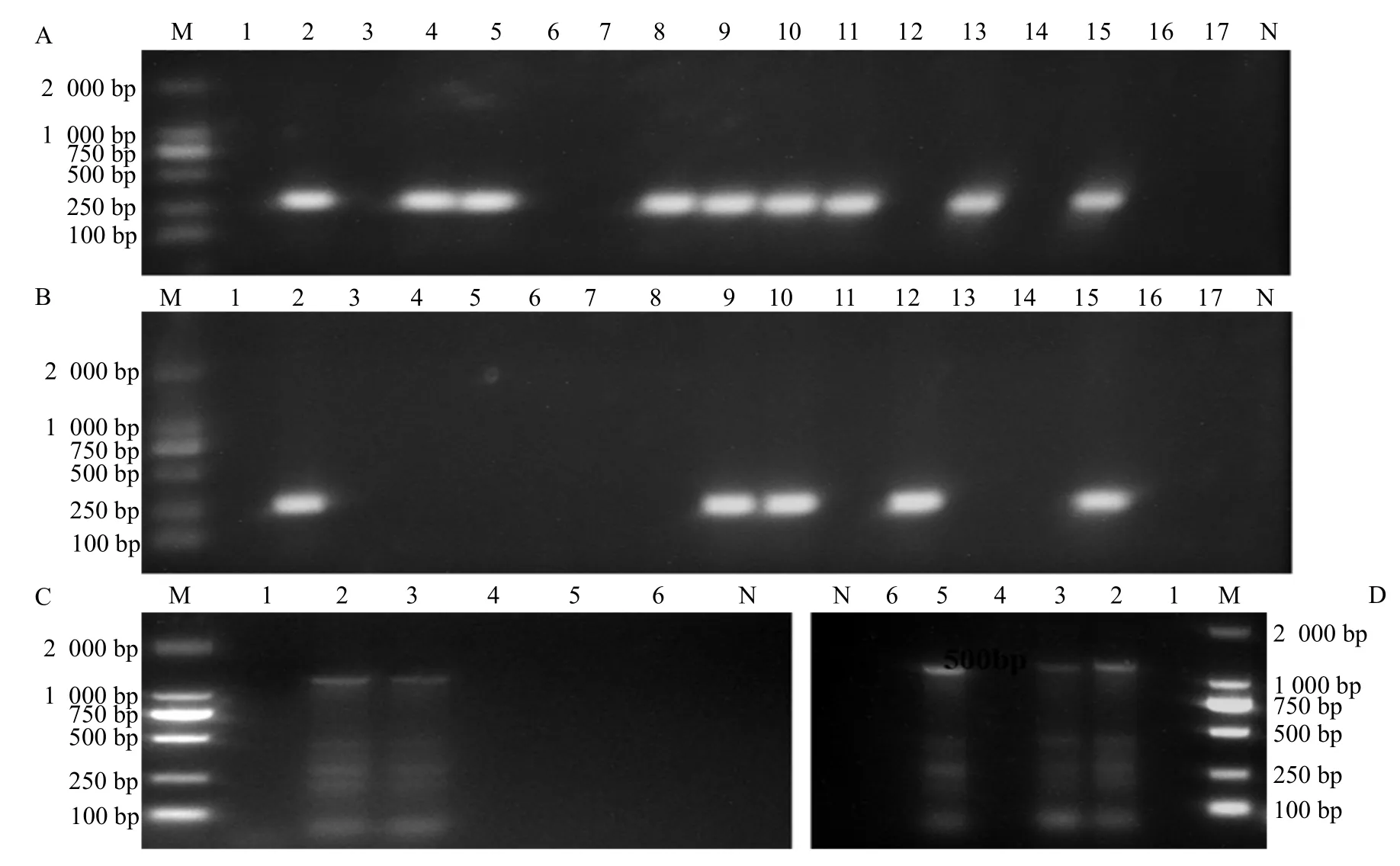
Fig. 3 PCR amplification of mcr-1 in DNA (A), plasmid (B), mcr-4 in DNA (C) and plasmid (D)
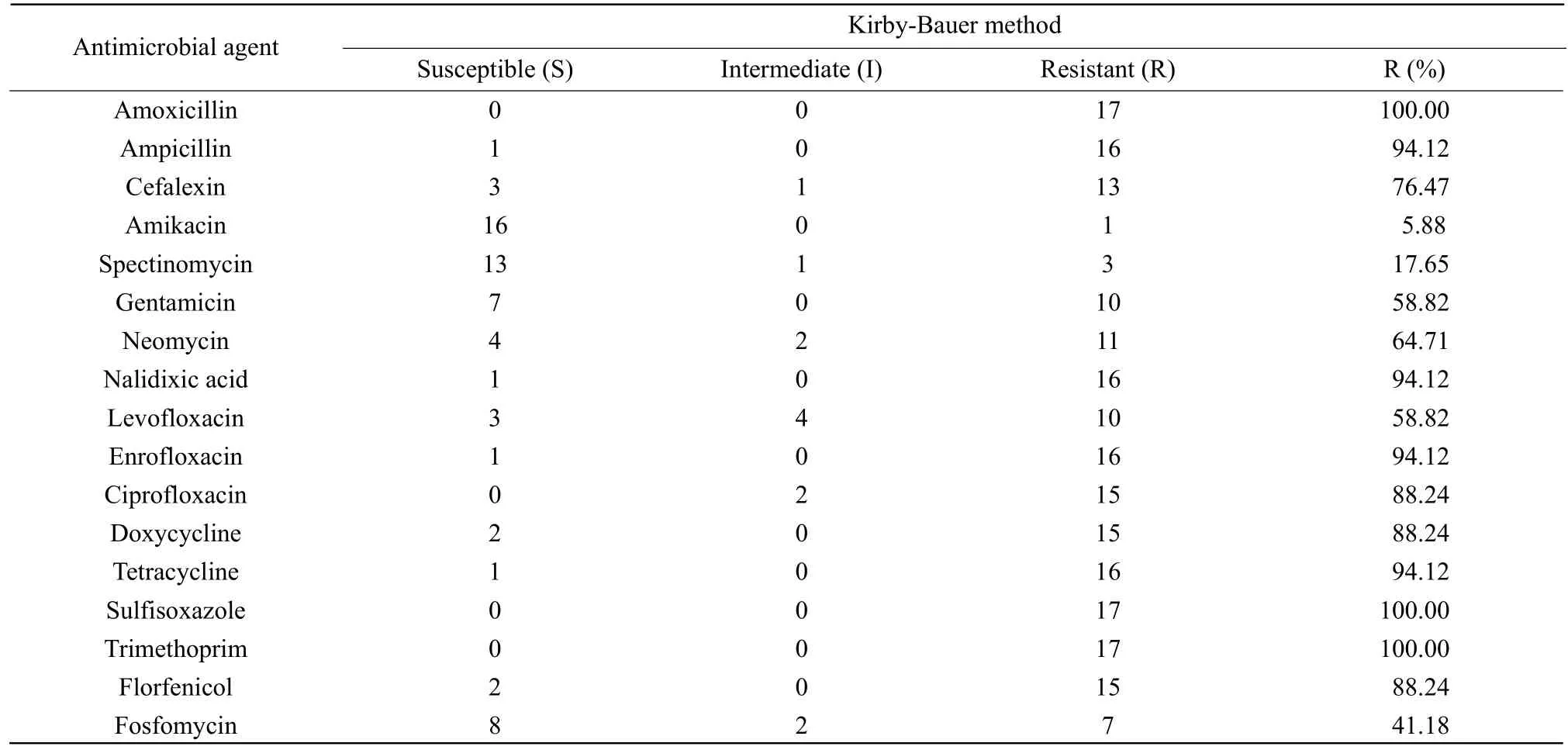
Table 4 Multiple resistance of 17 E. coli studied using K-B method
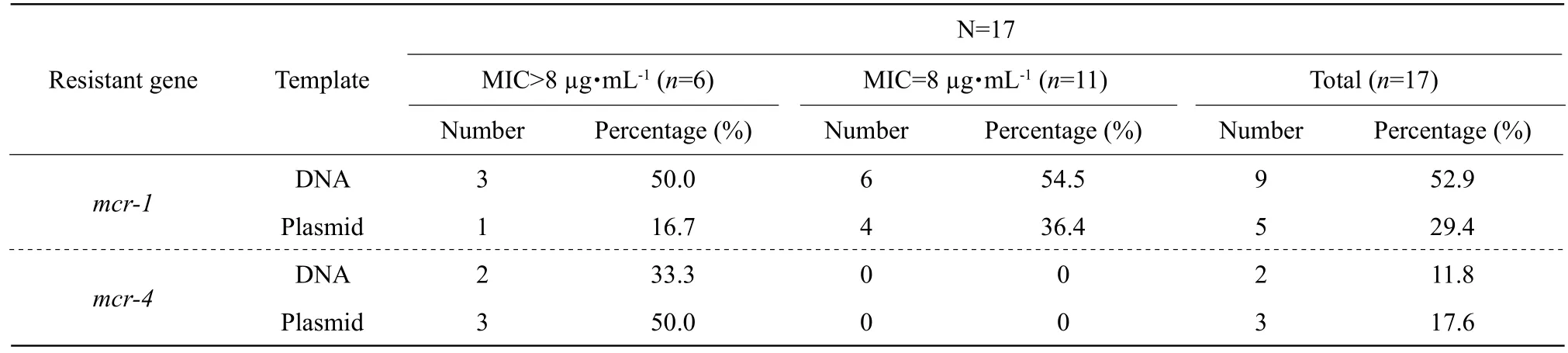
Table 5 Positive rate of mcr-1 and mcr-4 of 17 E. coli by PCR
Discussion
Bacterial infections remained the leading killer worldwide which was worsened by the continuous emergence of antibiotic resistance (Huet al., 2018).The rise in carbapenem resistance among gramnegative bacteria had renewed interest in colistin.This was a scenario where antimicrobials were used irrationally, and even in the propagation of livestock, poultry birds and for other veterinary purposes (Chikaet al., 2018). Recently, EUCASTCLSI Polymyxin Breakpoints Working Group declared that broth microdilution (BMD) was the only valid method for colistin susceptibility testing.Furthermore, despite the high error rates reported for disk and gradient diffusion methods, some laboratories continued to use these approaches due to the low cost and availability of necessary materials (Dafopoulouet al., 2015; Hindleret al., 2013). However, BMD methods were impractical for diagnostic laboratories owing to the considerable in-house preparation required, leading to high workload. For data accuracy,a total of 1 250 MICs of colistin were determined by BMD method in this study. MIC distribution could also been used in the process of setting clinical breakpoints if more accurate data were available. MIC value itself,however, should be interpreted with caution and was not directly translated to serum concentrations of antibiotics.
Due to insufficient data to establish colistin clinical breakpoints for Enterobacteriaceae, ECV was established by CLSI (2015) for certain Enterobacteriaceae based on MIC distribution data. EUCAST colistin susceptibility breakpoint of ≤2 μg • mL-1was used in 2017. ECV was set from a consideration ofin vitrosusceptibility data and as a result they could have no inherent clinical significance. However, Mirandaet al.(2016) argued that these ECVs did provide guidance to susceptibility testing laboratories as to how to report the meaning of the data they record. The preliminary ECV of colistin was 8 µg • mL-1among 1 250 MICs in this study. It was speculated that due to different amounts of colistin used in chickens in different regions, the results were not the same as those measured by CLSI. Maybe there was a temporary high breakpoint due to excessive use of colistin of China's farms. Drug sensitivity test results showed that the strain with low sensitivity to colistin was also less sensitive to most antibiotics. Interestingly,amikacin and spectinomycin were quite effective for these strains, but both of these were aminoglycosides that could be nephrotoxic. Intriguingly, colistin was closely related to its renal toxicity. Therefore, these two drugs were not probable to combine with colistin for antimicrobial infection.
The description of the plasmid-mediated colistin resistance genemcr-1 and its variants (mcr-2 to -8)were public health concerned given their potential to readily disseminate among clinical pathogens. Since,the discovery by Liuet al. (2016) of the plasmidborne phosphoethanol-amine transferase resistance determinantmcr-1 revealed a mechanism for horizontal spread. Until now,mcr-1 gene had been detected in multiple species of Enterobacteriaceae from both human and food producing animals in five continents and more than 40 countries (Mendeset al.,2018; Principeet al., 2018; Wanget al., 2017; Yiet al.,2017). Resistance to colistin inE. coliclinical isolates appeared low. In this study,mcr-1-positive isolates accounted for approximately 16.67% (1/6) of colistin resistantE. coliisolates in plasmid. Interestingly,mcr-4 gene was found to 50% (3/6). In addition,mcr-4 was positive only in colistin resistantE. coli. While,it was speculated thatmcr-1 mediated low level drug resistance.
Conclusions
A total of 1 250 MICs of colistin were determined by BMD method. MIC distribution was analyzed by ECOFF finder software, and the preliminary ECV of colistin was established (8 µg • mL-1). Accurate colistin susceptibility testing methods were necessary to ensure an optimal patient outcome and to reduce the spread of resistance. ECV facilitated drug resistance monitoring and provided scientific basis for the risk assessment of drug resistance. Besides, multiple resistance was studied by using K-B method. Furthermore, an increasing prevalence of multidrug-resistant gramnegative infections was found with limited treatment options. Nevertheless, further molecular researches are needed to study in depth the colistin resistant genesmcr-1 andmcr-4.
 Journal of Northeast Agricultural University(English Edition)2021年2期
Journal of Northeast Agricultural University(English Edition)2021年2期
- Journal of Northeast Agricultural University(English Edition)的其它文章
- Quercetin Increased Protein Utilization and Decreased Nitrogen Excretion in Broilers by Activating TOR Signaling Pathway
- Effects of Altitude Change on Nutritional Quality and Elymus nutans Regularity in Qinghai-Tibet Plateau
- Comprehensive Evaluation of Processing Quality of Tibetan Native Hulless Barley Variety by Factor Analysis
- Comparative Analysis of Bacillus thuringiensis Vip3Aa57 and Vip3Aa62 Insecticidal Activities
- Effects of Interaction of Soil Moisture and Organic Matter on Powdery Mildew Disease and Growth of Heracleum moellendorffii Hance
- Index Design and Comprehensive Evaluation of Germplasm Resources of Fruits Based on Mathematical Model of AHP and FCE: Sterculia nobilis Smith as a Case
