Quantitative Evaluation of the Larval Density of Sea Potato Acaudina spp. with Mitochondrial Molecular Marker
LI Xiang, WANG Guilin, LI Yunfan, LIU Wei, LU Chao,GUO Yuchen, HUANG Wen, WANG Jinhui, and DONG Bo, 2), *
Quantitative Evaluation of the Larval Density of Sea Potatospp. with Mitochondrial Molecular Marker
LI Xiang1), 2), #, WANG Guilin1), 2), #, LI Yunfan1), LIU Wei3), 4), LU Chao3), 4),GUO Yuchen3), 4), HUANG Wen3), 4), WANG Jinhui5), and DONG Bo1), 2), *
1),,,266003,2),,266237,3),352100,4),200137,5),,200137,
The sea potatospp. is a species of sea cucumber, belonging to the classunder Phylum. It becomes blooming recently in the East Sea of China, causing serious ecological problems. However, until now there are no molecular data for its larval identification and population genetic analysis. In this study, we firstly screened a mtDNA fragment and demonstrated that it was the species-specific molecular marker for the identification ofspp. We then developed a quantitative polymerase chain reaction (PCR)method to evaluate the larval density ofspp. based on this molecular probe. Utilizing this method, we examined 116 plankton samples collected in four seasons from 13 stations along the coastal region in Fujian province, China. The results showed that the high larval density was presented at stations 1, 2, and 3, which were near a quay in the coast. The larval density increased from April and reached the highest value in June and July, suggesting temperature might be the main environmental factor on the effects of its population distribution and density. Our work provides an important molecular tool for species identification and risk evaluation of a potentially invasive species.
spp.; sea potato; larval density; population distribution; molecular marker
1 Introduction
The sea potato (spp.), a species of sea cucum-ber,widely distributes in the waters alongsome Asiancounties including Philippine and China. Most of these spe- cies live in the intertidal zone, and only a few of them live in the coastal sediment. The sea potato has simple body structure with a fast reproductive ability. Every summer, it becomes blooming in the East Sea of China, causing serious problems on the maintenance and operation of marine facilities. For example, large numbers of adults and larvae of sea potato caused the blockage of the pipes for seawater intake and drainage for nuclear power plants. It is necessary to carry out species identification and establish the risk evaluation system forthis species. The classic species identification is mainly based on morphology observation. However, this approach is hard to identify the larvae of sea potato from a large number of plankton. The molecular methods based on the mitochondria genome have become an effective way to solve this problem.
Mitochondria are subcellular organelles unique to eukaryotes, playing an important role in a series of cytolo- gical processes, such as energy metabolism (Brand, 1997; Morenosánchez., 2010), cellular aging and diseases (Wallace., 1995). Mitochondrial genome data have been utilized to study phylogeographic (Scribner., 2003), evolution and phylogenetic relationships of the ani- mals in the metazoans (Boore., 2005). Mitochondrial DNA (mtDNA) is a double-stranded circular DNA molecule that is independent of the chromosome. Its size is about 16kb (Boore, 1999). Except for a few species, most of animals’ mitochondrial genomes consist of 37 genes: 13 protein-coding genes, 2 ribosomal RNAs (rRNAs), 22 tran- sfers RNAs (tRNAs) (Boore and Brown, 1994; Peregrino- Uriarte., 2009), and a non-coding putative control re- gion, in which signal sequences exist for transcription and replication (Takata., 2001). mtDNA is essential for the process of protein synthesis, transcription, and translation. In addition, it has the characteristics of the small genome (Behera., 2018), compact structure, high copy number (Ingman., 2000) and no rearrangement during cell meiosis (Fan., 2011). It is widely used in evolution analysis, genetic diversity study and species identification (Curole and Kocher, 1999). The full mitochondrial sequ- ences of most species of the sea cucumber have been de- termined (Shen., 2009; Perseke., 2010).
In our previous study, the mitochondrial DNA sequence ofspp.has been detected through polymerase chain reaction (PCR) amplification and Sanger sequencing (Wang., 2019). In this study, based on the complete mitochondrial genome sequences, we screened and identi- fied the partial sequence ofgene as a specific molecular marker for the identificationofspp. from the col- lected plankton samples. With the molecular marker, we de- veloped an effective method to identify and evaluate the population dynamic ofspp. through PCR.
2 Materials and Methods
2.1 Animal Collection
Four adults ofspp.(Fig.1A) were collected fromthe coast in Fujian Province, China. Species were identifiedby morphology (Xiao, 2015) and 16S rRNA sequencing based on the published sequences (Wen., 2011).The samples were then fixed in 75% ethanol and stored at 4℃ until DNA extraction.
The larvae ofspp.were collected using a type II plankton collection net (diameter 31.6cm, length 140cm,mesh size 0.16mm, rope length 10m) from 13 stations. The plankton samples were concentrated in a 500mL plastic sampling bottle and then fixed with 75% ethanol. The po- sitions of sampling stations were indicated in Fig.3A.
2.2 DNA Isolation
Total genomic DNA was extracted from the gonad of adultspp. and the planktonic samples, respec-tively, using a modified phenol/chloroform/isoamyl alco- hol method (Wei., 2020). Firstly, 45mL STE buffer (100mmolL−1NaCl, 10mmolL−1Tris-HCl, 100mmolL−1EDTA, pH 8.0) and 4.5mL 10% SDS (dissolved in ddH2O) were mixed in a 50mL tube to make the lysis buffer. The gonads were dissected from the animal and were put into a 1.5mL tube. Then 700μL lysis buffer and 2.8μL Protei- nase K (Merck, dissolve in ddH2O with the concentration of 50mgmL−1) were added into the tube to make a final concentration of 200μgmL−1proteinase K. The mixture was shaken gently and incubated in a water bath at 58℃for 3h.Finally, DNA was extracted with phenol-chloro- form-isoamyl alcohol and chloroform-isoamyl alcohol, re- spectively, and then precipitated by alcohol and dissolved in ddH2O. The dissolved DNA was stored at −20℃.
2.3 PCR Amplification
PCR reaction was carried out using PfuS DNA polyme- rase (gift from Dr. Zhiyi Lv) in a 50μL volume. The reac- tion solution includes 10μL 5× Phusion HF Buffer (Ther- mo fisher Catalog number: F518L), 1μL 10mmolL−1dNTP, 2.5μL 10μmolL−1Primer F, 2.5μL 10μmolL−1Primer R, 1μL template DNA (200ngμL), 0.5μL PfuS DNA poly- merase, and 32.5μL ddH2O. The PCR was performed as follows: Pre-denaturation at 95℃ for 3min; denaturation at 95℃ for 15s; annealing at 55℃ for 15s, followed withelongation at 72℃ for 1min, and totally for 35 cycles; and a final extension at 72℃for 5min. The PCR products were purified by Gene JET Gel Extraction Kit (Thermo Fisher Scientific, Lithuanian).

Table 1 Primer sequences and annealing temperatures
Note: Degenerate base: R=A/G, Y=C/T, H=A/T/C, D=G/A/T.
The primers for the amplification offromspp.were designed based on the published mitoge- nomes (Wang., 2019).The primer sequences and an- nealing temperaturesof PCR were shown in Table 1.
2.4 TOPO Clone, Sequencing, and Sequence Alignment
TOPO cloning was performed using pEasy-Blunt3 kit(Transgen, Beijing, China). Firstly, 4μL purified PCR pro- duct was mixed with 1μL pEasy-Blunt3 vector and incubated at 25℃ for 15min.Then they were transformed intoTrans1-T1 competent cells and spray on Luria-Bertani (LB)agar plates. The LB plates were cultured overnight at 37℃. Monoclonal colonies were picked up for sequencing. Sequencing was performed by Genwiz company (Tianjin, Chi- na). Multiple sequence alignments were carried out using the ClustalW program in BioEdit software (version 7.0.4.1) with default parameters.
2.5 Image Analysis and Statistical Analyses
Totally 116 plankton samples were examined by PCRto determine whether the larvae ofsppwere in the samples. PCR products were detected on a 1% agarose gel.The electrophoresis bands of 116 samples were analyzed using ImageJ software to calculate the gray value, which can reflect the content of DNA.(also known asgene) was used as a reference. The distribution ofspp. larvae at 13 detected stations were eva- luated based on molecular data.
3 Results
3.1 Morphological and Molecular Identification of Acaudina spp.
One adult sample was collected from the coast of Fujian province. It was about eight cm long and its body pre- sented the light brown color with dark brown spots (Fig.1A). They were morphologically similar to sea potato. To iden- tify the species, we designed primersandto amplify a mitochondrial fragment including two genesandbased onpreviously published sequ- ences (FJ971405 and FJ971380) ofspp.(Wen., 2011)A 1500 bp DNA fragment was amplified from the total DNA (Fig.1B). After sequencing and alignment,bothandgenes from our samples showed high identify with the ones fromspp..However, the identity ofsequence from our samples with FJ- 971405 was only 85% (Fig.1C), and thesequ- ence from our samples showed 90% identity with FJ971- 380 (Fig.1D). Based on these data, we identified our sam- ples asspp.
3.2 The Fragment of Mitochondrialnd1 Gene Was an Effective Species-Specific Molecular Marker
Mitochondrial genes are widely used in population iden- tification and genetic studies, such as(Das., 2018),(Wang., 2018),(Behera., 2015)In our previous work, we got the mitochondrial genome sequ- ence ofspp.. Based on this data, we designed pri- mers to amplify different fragments from the total DNA ofspp. and other marine species to identify the species-specific marker. The results showed that a 229bpDNA fragment ofgene could be effectively amplified through a primers pair,(Fig.2A) and(Fig.2B) from the total DNA ofspp., but could not from other marine species including ascidians (,), clam ()andscallop ()(Fig.2C). We then tested the validity of this pair of primers using total DNA extracted from the collected plankton samples. The results showed that one specific fragment could be successfully amplified from 9 of 13 samples (Fig.2D), suggesting that the fragment ofgene is an effective species-specific molecular marker for species identification and larva detection from the plankton samples.

Fig.1 Morphological and molecular identification of sea potato Acaudina spp.(A) Image of adultAcaudina spp.. Bar is 1cm. (B) Polymerase chain reaction (PCR) product with about 1500bp size. The length of the markers from top to bottom is 5000bp, 3000bp, 2000bp, 1500bp, 1000bp, 750bp, 500bp, 250bp, and 100bp, respectively. (C)Sequence alignment be- tween amplicon and Am-COI (FJ971405). The identity is 85%. (D) Sequence alignment between amplicon and Am-16S(FJ971380). The identity is 90%.

Fig.2 Screen of species-specific molecular for the identification of Acaudina spp. (A) Alignment of the forward primer with the nd1 sequences from different species. (B) Alignment of the reverse primers with the nd1 sequences from different species. (C) The amplificated bands were presented in 1% agarose gel using the nd1 gene primers and the total DNA from different marine species. M, DL5000 marker; 1, C. robusta, 2, S. clava, 3, Acaudina spp.,4, M. quadrangularis, 5, P. yes- soensis. (D) The amplificated bands were presented in 1% agarose gel using the nd1 gene primers and the total DNA from the nine collected plankton samples.
3.3 Quantitative Evaluation of Larval Population of Sea Potato
To evaluate the population of sea potato, we developed a relative quantitative analysis method based on the species-specificmarker. The quality of DNA from each sample was verified bywith universal primers.The optical density ratio ofandPCR bands were used to represent the relative quantity of sea potato larvae, and compare the population variation among dif- ferent samples. The 116 samples were collected from 13 stations distributed along the coast region of Fujian province (Fig.3A). We divided all the sampling stations into four groups by the distance to the S01 station: group I in- cluded S01, S02, and S03, which were off S01 station less than 1km; group II included S04, S05, S06 and S07, off S01 station 1to 2km; Group III included S08, S09, and S10, off S01 station 4 to 6km; group IV included S11, S12, and S13, off S01 station more than 9km. The total DNA was extracted from the samples, respectively. AllDNA samples were diluted to 20ngμL−1, and 10μL was utilized for PCR. The DNA quality of most samples (107/ 116) were sufficiently good for the amplification of the fragments ofand. The results showed ob- vious differences in the relative larval density among the samples and the groups (Fig.3B). It clearly showed that the high-density sea potato was presented in group I station in spring and summer near the land, suggesting that distribution of sea potato is associated to the marine se- diment environment.
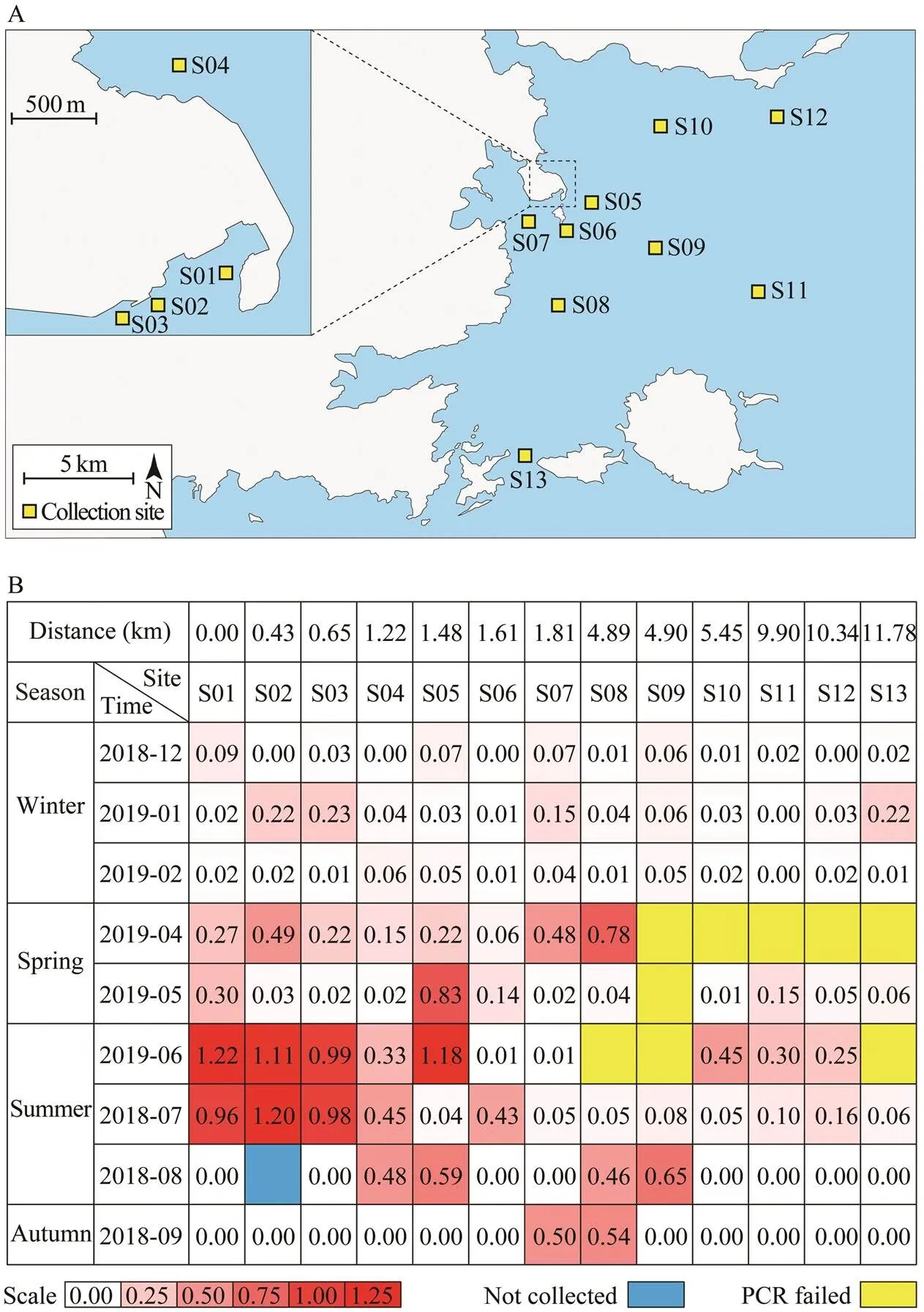
Fig.3 Quantitative evaluation of the density of Acaudina spp. larvae. (A) Sampling stations in this study (B) Relative quan- tity of Acaudina spp. larval density.Relative densities of Acaudina spp. larvae are indicated through a gradient change from white to red. White color indicates lower density of larvae. Red color indicates higher density of the larvae. Blue color indicates that the sample was not collected. Yellow color indicates failures of PCR reaction (no band on the gel).
3.4 Season Variation of Acaudina spp.Larvae
The samples were collected in four seasons. The data showed that the highest larval density appeared in June and July. For stations in group I, there was a significant increase from May to July, and then a rapid decline after September (Fig.4A). For stations in groups II and III, they showed similar patterns: the lowest density in Winter, and other seasons showed the moderate density (Figs.4B and 4C). For group IV, the relative density was globally low and two small peaks were observed in January and June (Fig.4D).
4 Discussion
In this study, we successfully identified a 229bp fragment of geneasspp. specific molecular mar- ker, which provides an effective molecular tool for species identification and population dynamic evaluation. By analyzing the relative quantity of this fragment with PCR, we quantitatively detected the larval density ofspp.in a marine coast, and found that their density decreased gradually from offshore to deeper waters. In the samples that were collected from about 20m deep of the seawaters, the larval density is quite low. In addition, the higher density of larvae was presented in the samples col- lected in May, June, and July, suggesting that the larvae ofspp. might propagate from April to July.
It is worthy to note that the method that we developed in this studycannot distinguish the species within one ge- nus. CRISPR-based specific high sensitivity enzymatic re-porter unlocking technology (Gootenberg., 2017; Gootenberg., 2018) and DNA endonuclease targeted CRISPR trans reporter technology (Chen., 2018) can be the alternative ways to accurately quantify target mole- cules. Loop-mediated isothermal amplification is another potentially useful technology with high specificity and ef- ficiency under isothermal conditions (Notomi., 2015).
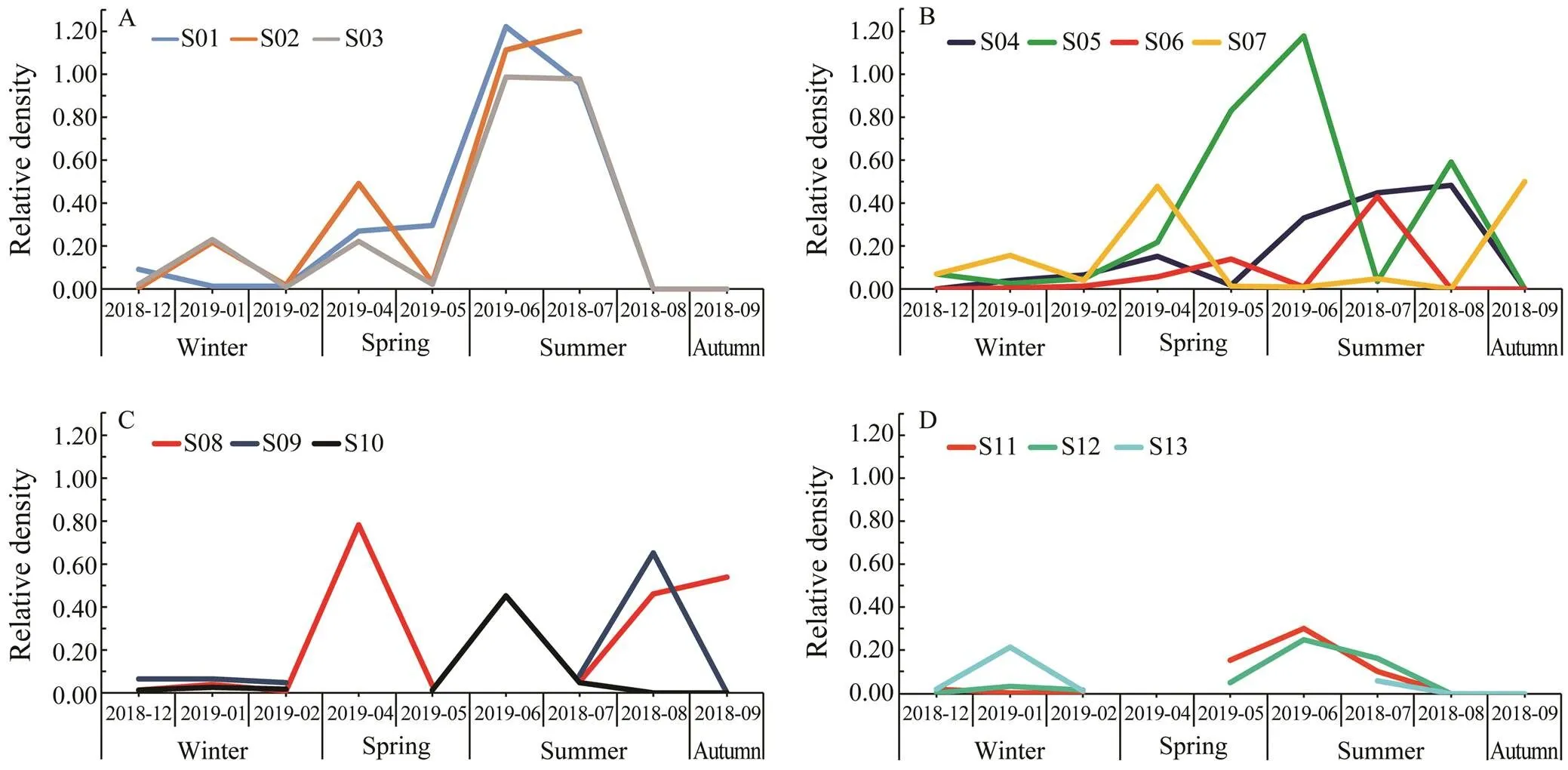
Fig.4 Relative density of Acaudina spp. larvae at different stations. Variation trend of relative density plot in stations of group I (A), group II (B), group III (C), and group IV (D), respectively.
Our results showed that the larval density ofspp. is dynamic and tightly correlates with the temperature and the sediment in the offshore seawaters. These re- sults therefore provide useful information to understand the seasonal and regional distribution ofspp. lar- vae. Based on this information, population dynamics can be effectively evaluated, early warning mechanisms canbe established, and the population density ofspp.can be regulated in the specific sea region.
Acknowledgements
This work was supported by the National Key Research and Development Program of China (No. 2018YFD090 0705), the Key Laboratory of Integrated Marine Monitor- ing and Applied Technologies for Harmful Algal Blooms Funds (No. MATHAB201706), and the Fundamental Re- search Funds for the Central Universities (No. 201822016).
Behera, B. K., Baisvar, V. S., Rout, A. K., Pakrashi, S., Kumari, K., Panda, D., Das, P., Parida, P. K., Meena, D. K., Bhakta, D., Das, B. K., and Jena, J., 2018. The population structure and genetic divergence of(Hamilton, 1822) analyz- ed through mitochondrial DNA cytochrome b gene for con- servation in Indian waters., 29: 543-551.
Behera, B. K., Kunal, S. P., Paria, P., Das, P., Meena, D. K., Pa- krashi, S., Sahoo, A. K., Panda, D., Jena, J., and Sharma, A. P., 2015. Genetic differentiation in Indian Major Carp,(Hamilton, 1822) from Indian Rivers, as revealed by direct sequencing analysis of mitochondrial Cytochromere- gion., 26: 1-3.
Boore, J. L., 1999. Animal mitochondrial genomes., 27: 1767-1780.
Boore, J. L., and Brown, W. M., 1994. Mitochondrial genomes and the phylogeny of molluscs.,108 (supp. 2): 61- 78.
Boore, J. L., Macey, J. R., and Medina, M., 2005. Sequencing andcomparing whole mitochondrial genomes of animals., 395: 311.
Brand, M. D., 1997. Regulation analysis of energy metabolism., 200: 193-202.
Chen, J. S., Ma, E., Harrington, L. B., Da Costa, M., Tian, X., Palefsky, J. M., and Doudna, J. A., 2018. CRISPR-Cas12a tar- get binding unleashes indiscriminate single-stranded DNase activity., 360: 436.
Curole, J. P., and Kocher, T. D., 1999. Mitogenomics: Digging deeper with complete mitochondrial genomes., 14: 394-398.
Das, S. P., Swain, S., Jena, J., and Das, P., 2018. Genetic di- versity and population structure ofreveal- ed by mitochondrial ATPase 6 gene., 29: 495-500.
Fan, S., Hu, C., Wen, J., and Zhang, L., 2011. Characterization of mitochondrial genome of sea cucumber: A novel gene arrangement in Holothuroidea., 54: 434-441.
Gootenberg, J. S., Abudayyeh, O. O., Kellner, M. J., Joung, J., Collins, J. J., and Zhang, F., 2018. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6., 360: 439.
Gootenberg, J. S., Abudayyeh, O. O., Lee, J. W., Essletzbichler, P., Dy, A. J., Joung, J., Verdine, V., Donghia, N., Daringer, N. M., Freije, C. A., Myhrvold, C., Bhattacharyya, R. P., Livny, J., Regev, A., Koonin, E. V., Hung, D. T., Sabeti, P. C., Col- lins, J. J., and Zhang, F., 2017. Nucleic acid detection with CRISPR-Cas13a/C2c2., 356: 438.
Ingman, M., Kaessmann, H., Pääbo, S., and Gyllensten, U., 2000. Mitochondrial genome variation and the origin of modern hu- mans., 408: 708-713.
Morenosánchez, R., Rodríguezenríquez, S., Marínhernández, A., and Saavedra, E., 2010. Energy metabolism in tumor cells., 274: 1393-1418.
Notomi, T., Mori, Y., Tomita, N., and Kanda, H., 2015. Loop- mediated isothermal amplification (LAMP): Principle, features, and future prospects., 53: 1-5.
Peregrino-Uriarte, A. B., Varela-Romero, A., Muhlia-Almazan, A., Anduro-Corona, I., Vega-Heredia, S., Gutierrez-Millan, L. E., De la Rosa-Velez, J., and Yepiz-Plascencia, G., 2009. The complete mitochondrial genomes of the yellowleg shrimpand the blue shrimp(Crustacea: Decapoda)., 4: 45-53.
Perseke, M., Bernhard, D., Fritzsch, G., Brümmer, F., Stadler, P. F., and Schlegel, M., 2010. Mitochondrial genome evolutionin Ophiuroidea, Echinoidea, and Holothuroidea: Insights in phy- logenetic relationships of Echinodermata., 56: 201.
Scribner, K. T., Talbot, S. L., Pearce, J. M., Pierson, B. J., Bol- linger, K. S., and Derksen, D. V., 2003. Phylogeography of Canada Geese () in western North America., 120: 889-907.
Shen, X., Tian, M., Liu, Z., Cheng, H., Tan, J., Meng, X., and Ren, J., 2009. Complete mitochondrial genome of the sea cu- cumber(Echinodermata: Holothuroi- dea): The first representative from the subclass Aspidochi- rotacea with the echinoderm ground pattern., 439: 79- 86.
Takata, K., Yoshida, H., Hirose, F., Yamaguchi, M., Kai, M., Oshige, M., Sakimoto, I., Koiwai, O., and Sakaguchi, K., 2001.mitochondrial transcription factor A: Characteri- zation of its cDNA and expression pattern during development., 287: 474-483.
Wallace, D. C., Shoffner, J. M., Trounce, I., Brown, M. D., Bal- linger, S. W., Corraldebrinski, M., Horton, T., Jun, A. S., and Lott, M. T., 1995. Mitochondrial DNA mutations in human de- generative diseases and aging., 1271: 141-151.
Wang, G., Li, X., Wang, J., Zhang, J., Liu, W., Lu, C., Guo, Y., and Dong, B., 2019. The complete mitochondrial genome and phylogenetic analysis of.–, 4: 668-669.
Wang, X., Han, X., Zhang, Y., Liu, S., and Lin, Q., 2018. Phy- logenetic analysis and genetic structure of the seahorse,from the Arabian and Red Sea based on mi- tochondrial DNA sequences., 39: 165-171.
Wei, J., Zhang, J., Lu, Q., Ren, P., Guo, X., Wang, J., Li, X., Chang, Y., Duan, S., Wang, S., Yu, H., Zhang, X., Yang, X., Gao, H., and Dong, B., 2020. Genomic basis of environmen- tal adaptation in the leathery sea squirt ()., 20: 1414-1431.
Wen, J., Hu, C., Zhang, L., and Fan, S., 2011. Genetic identifi- cation of global commercial sea cucumber species on the basis of mitochondrial DNA sequences., 22: 72-77.
Xiao, N., 2015.. Science Press, Beijing, 100pp (in Chinese).
June 6, 2020;
September 21, 2020;
November 17, 2020
© Ocean University of China, Science Press and Springer-Verlag GmbH Germany 2021
#The two authors contributed equally to this work.
. E-mail: bodong@ouc.edu.cn
(Edited by Qiu Yantao)
 Journal of Ocean University of China2021年3期
Journal of Ocean University of China2021年3期
- Journal of Ocean University of China的其它文章
- Effects of Nitrogen Sources and Concentrations on the Growth of Different Phytoplankton Taxa
- Mathematical Proof of the Synthetic Running Correlation Coefficient and Its Ability to Reflect Temporal Variations in Correlation
- Risk Assessment of Marine Environments Along the South China Sea and North Indian Ocean on the Basis of a Weighted Bayesian Network
- Assessment of the Tidal Current Energy Resources and the Hydrodynamic Impacts of Energy Extraction at the PuHu Channel in Zhoushan Archipelago, China
- Changes in the Photosynthetic Pigment Contents and Transcription Levels of Phycoerythrin-Related Genes in Three Gracilariopsis lemaneiformis Strains Under Different Light Intensities
- Influence of Environmental Conditions on the Sound Velocity Ratio of Seafloor Surficial Sediment
