First molecular cytogenetic characterisation of tracheal squamous cell carcinoma cell line KLN 205
Shaymaa Azawi, Thomas Liehr, Martina Rincic2Institute of Human Genetics, Jena University Hospital, Friedrich Schiller University, Jena 07747, Germany.
2Croatian Institute for Brain Research, School of Medicine University of Zagreb, Zagreb 10000, Croatia.
Abstract
Keywords: Lung cancer, squamous cell carcinoma (SCC), murine cell line KLN 205, murine multicolour banding(mcb), array comparative genomic hybridisation (aCGH)
INTRODUCTION
Among the leading adverse forms of acquired malignant diseases is human lung cancer representing ~17%of new cancer diagnoses worldwide. About 30% of lung-associated cancers are of squamous cell carcinoma(SCC) type[1], and it has been shown that 80%-90% of lung SCCs (L-SCCs) are associated with tobacco consumption, i.e., smoking[1,2]. Being a long-standing clinical problem, there are numerous therapeutic options at hand, such as carboplatin- or cisplatin-based cytotoxic chemotherapy, EGFR targeting therapy(e.g., by erlotinibin or cetuximab), anti-angiogenesis treatment by ramucirumab or approaches targeting the immune system with specific inhibitors, e.g., ipilimumab or nivolumab[1]. However, not all L-SCC patients can be treated optimally using the currently available options, thus research on, e.g., how to access the FGFR[3], IGF[4]or PI3K-Akt/mTOR pathways[5], is ongoing.
Murine cancer cell lines are well established models to perform such studies[3-5]. However, most of these murine cancer cell lines have never been (cyto)genetically characterised in detail, irrespective of how often they have already been used in research[6,7]. This is because murine chromosomes are hard to distinguish in banding cytogenetics[7].
Thus, the murine cell line KLN 205 (also referred to as KLN205 or KLN-205), being derived from a chemically induced tracheal SCC in 1978[8], has also never been characterised using cytogenetics, molecular cytogenetics or molecular karyotyping, even though it has been applied in almost 100 studies. Only the modal chromosome number was determined as being between 73 and 76[9]. The cell line was induced in the lung of a maleDBA/2mouse by repeated intra-tracheal injections of 3-methylcholanthrene. Then, the tissue was serially passaged inDBA/2mice intramuscularly, and the 9th and 10th transplant generations were further used as “Nettenheim carcinoma”[10], known to be composed of heterogeneous cell populations[9].Nettenheim carcinoma has been classified as tracheal SCC, as well as been called lung carcinoma, from which the cell line KLN 205 was established by serial sub-cultivation and subcloning[10]. Interestingly, the American Type Culture Collection provides the cell line as “forming metastatic lesions in lungs after inoculation into mice” and “SCC cell line”[11]. Overall, there seems to be some confusion regarding for which human tumour KLN 205 cells can be used as models. Accordingly, the KLN 205 cell line has been used in studies considering, for example, the following types of cancer:
• SCC[12-14]
• Lung cancer[15-22]
• Non-small cell lung cancer[23]
• L-SCC[24,25]
• Lung-metastatic tumour cells[26]
• Tongue cancer[27]• Subcutaneous SCC[28]
It was shown previously that a comprehensive molecular cytogenetic characterisation of murine tumour cell lines can define the cancer subtype to which they belong because the cytogenomic profile of chromosomal imbalances is typically specific to a certain tumour type[6].
Thus, the karyotype, a comprehensive map of chromosomal imbalances, and anin silicotranslation of these results to the human genome were performed here for the KLN 205 cell line to possibly determine to which human cancer type it is most similar.
METHODS
Cell line
The murine KLN 205 cell line (American Type Culture Collection, ATCC CRL-1453™; Wesel Germany) was grown adherently according to the recommendations of the provider. The cells were cytogenetically worked up[28]and the whole genomic DNA was extracted from the same passage of cells[29]. Molecular cytogenetic and molecular karyotyping [array comparative genomic hybridisation (aCGH)] analyses were done as outlined below.
The KLN 205 cell line was specifically purchased for this study from ATCC. The provider guaranteed the identity of the cell line.
Molecular cytogenetics and karyotyping
Fluorescencein situhybridisation (FISH) was performed as previously described[29]using whole chromosome paints (“SkyPaintTM DNA Kit M-10 for Mouse Chromosomes”, Applied Spectral Imaging,Edingen-Neckarhausen, Germany) for multicolour-FISH (mFISH) and murine chromosome-specific multicolour banding (mcb) probe mixes for FISH-banding[29]. At least 30 metaphases were analysed for each probe set Zeiss Axioplan microscopy, equipped with ISIS software (MetaSystems, Altlussheim, Germany).aCGH was done according to standard procedures by “SurePrint G3 Mouse CGH Microarray, 4×180K”(Agilent Technologies)[29].
Data analyses
Imbalances and breakpoints of KLN 205 were determined according to mcb and aCGH data and aligned to human homologous regions using Ensembl and the UCSC Genome Browser, as previously described[29]. The obtained data were compared to the known genetic changes in human cancers[30-32].
RESULTS
FISH and aCGH
The approximately tetraploid cell line KLN 205 had a relatively stable karyotype, falling into four well defined clones with minor but stable differences: Clone 1 (43.3%) [Figure 1], 77-82,XXYYYY,-2,del(3)(A3D),der(3)(A1→A3::D→G3:),-4,+del(5)(C3),-7,-8,der(9)t(9;10)(A5;A1),-13,der(14)t(2;14)(A2;A1),+15,-17,del(19)(D); Clone 2 (20%), same as Clone 1, but +del(5)(C3) instead of +5 and four chromosome 12 instead of three; Clone 3 (13.4%), same as Clone 2 but four chromosome 2; and Clone 4 (13.4%), same as Clone 2 but four chromosome 2, no der(9)t(9;10)(A5;A1).
Overall, the FISH results are in agreement with those of the aCGH analysis, as summarised in Figure 2A. Anin silicotranslation of those results to the human genome (only imbalances larger than 3.5 megabase pairs were included in the evaluation) identified the corresponding homologous region in the human genome[Figure 2B]. The genomic details are presented in Supplementary Table 1. The aCGH results for murine Ychromosome are not informative.
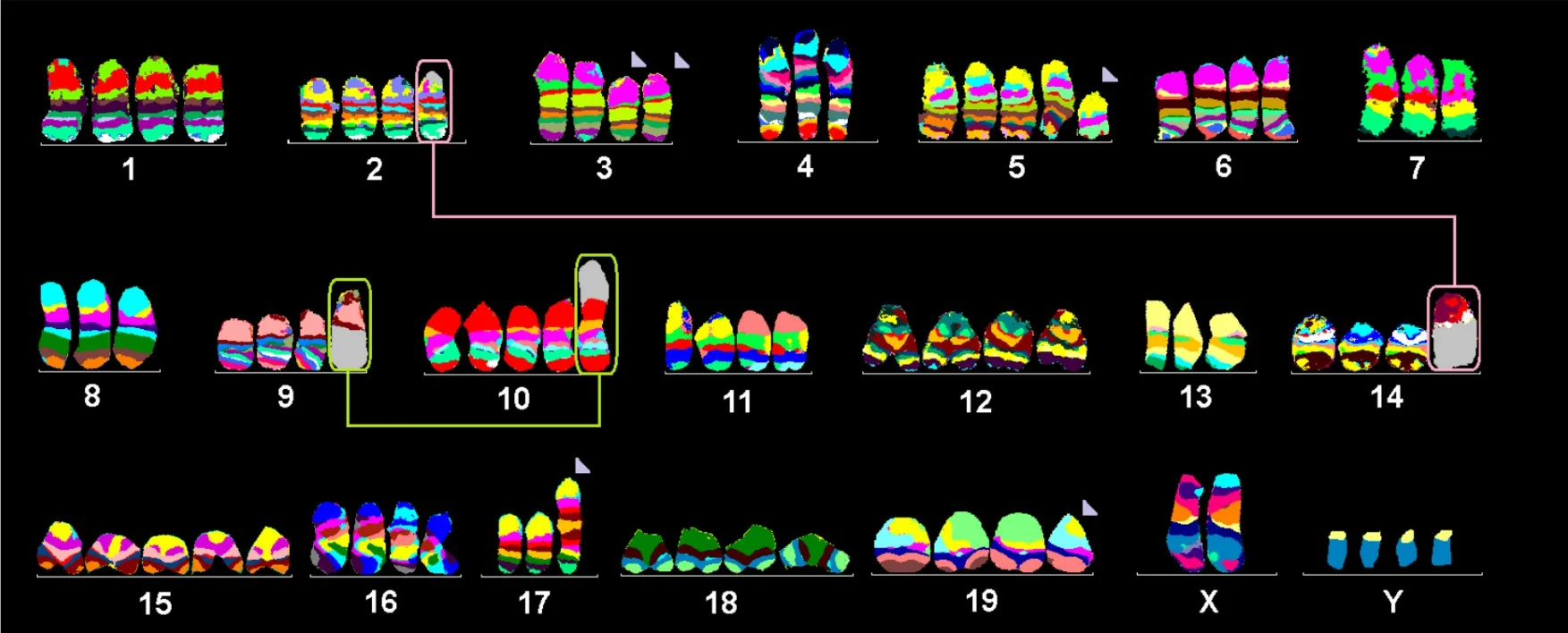
Figure 1. Murine multicolour banding (mcb) was applied on chromosomes of the KLN 205 cell line. This figure depicts the summary of 20 chromosome-specific FISH experiments as typical pseudo-colour banding. Derivative chromosomes consisting of different chromosomes are highlighted by frames and shown twice in this summarising karyogram.
Comparison with literature
The corresponding translated homologous CNV regions of the KLN 205 cell line [Figure 2B] were compared with the common imbalances in related human cancers[30-32], as summarised in Table 1. In human non-SCC (excluding small cell lung cancers as well), only 7/28 regions (25%) were affected by copy number variations (CNVs) as in the KLN 205 cell line. Surprisingly, the three other compared human cancer subtypes [human lung cancer (SCC-type), human small cell lung cancer type and head and neck cancer(SCC-type = HNSCC)] revealed CNVs in 12 regions (~43%) each that are in the KLN 205 cell line.

Figure 2. aCGH results and copy number variations detected in the KLN 205 cell line are summarised here with respect to a diploidbasic karyotype. Gains are shown as green bars, losses are shown as red bars and breaks are registered as arrows. (A) The imbalances found in the cell line depicted along a murine chromosome set. (B) The results translated and projected along the human chromosome set.
DISCUSSION
The KLN 205 cell line has been applied in around 100 published research studies. While morphologically it seems non-negotiable that KLN 205 in culture appears similar to SCC cells[12-14,24,25], they have been applied as models for lung cancer in general[15-22], metastatic lung cancer cells[26]and even non-small cell lung cancer[23]. Surprisingly, they have even worked as a model for tongue cancer[27]and subcutaneous SCC[28].
The present study showed that KLN 205 had an overall stable, approximately tetraploid karyotype with 77-82 chromosomes per cell. Thus, this > 40-year-old cell line still has about the same chromosome number as reported in 1980[9]. Tetraploidisation was found to obviously have been an early event, possibly already being present in the original tumour or acquired during cell line establishment and early adaptation of the cells to culture conditions[6]. Structural chromosomal aberrations are in the lower range of tumour cell lines(see, e.g., the NIH3T3 line[7]). Thus, the use of KLN 205 as a model for metastatic cancer cells[26]is not justified based on the chromosomal aberrations.
Reviewing the literature, SCCs, independent of their localisation, show relatively few differences in terms of acquired CNVs. It has been stated for HNSCCs that they have a rather uniform CNV pattern, irrespective of whether they derive, e.g., from the larynx, oesophagus or tongue[33]. The same was found when comparing HNSCCs and L-SCCs in this study [Table 1]. In addition, it has been shown that the patterns of gains and losses in human lung cancer (SCC-type) and human small cell lung cancer type are strikingly similar as well[31]. Thus, their use as models for SCC cells in general[12-14,24,25]or even for subcutaneous SCC[28]seems to be justifiable. In addition, considering their genetic background, KLN 205 can use as a model for lung cancer in general[15-22], as as well as for even non-small cell lung cancer[23]. Their use as a tongue cancer model[27]would only be appropriate if SCC is the cancer type being assessed.

Table 1. Copy number changes associated with molecular subtypes of human lung cancer (SCC-type), human lung cancer (non-SSCtype and non-small cell lung cancer-type), human small cell lung cancer and head and neck cancer (SCC-type = HNSCC), as shown in[30-32], compared with the copy number variants (CNVs) in the KLN 205 cell line. Concordances with human CNVs are highlighted in bold
In conclusion, according to the genetic profile of tetraploid KLN 205, this cell line is primarily a model for human SCC. It can also be used to study lung cancer, not restricted to L-SCC, due to the general similarities of CNVs in lung cancer in general. This study may be the starting point for further genetic characterisation of this interesting cell line by other cytogenomic approaches, including second-generation sequencing.
DECLARATIONS
Acknowledgments
The technical support of Nadezda Kosyakova (Jena, Germany) is kindly acknowledged.
Authors’ contributions
Developed the idea for the study and got funded for it: Liehr T
Did the FISH-studies: Azawi S
Performed aCGH studies and did pre-evaluation: Rincic M
Performed the overall data interpretation: Azawi S
Did final paper drafting: Liehr T, Azawi S
All authors agreed on final draft.
Availability of data and materials
Not applicable.
Financial support and sponsorship
This work was supported by grant # 2013.032.1 of the Wilhelm Sander-Stiftung.
Conflicts of interest
All authors declared that there are no conflicts of interest.
Ethical approval and consent to participate
According to the ethical committee (medical faculty) and the Animal Experimentation Commission of the Friedrich Schiller University, there are no ethical agreements necessary for studies involving murine tumour cell lines such as KLN 205.
Consent for publication
Not applicable.
Copyright
© The Author(s) 2021.
 Journal of Cancer Metastasis and Treatment2021年7期
Journal of Cancer Metastasis and Treatment2021年7期
- Journal of Cancer Metastasis and Treatment的其它文章
- GENERAL INFORMATION
- Covid19 and cancer care: the end of the beginning
- Adjuvant therapy for renal cell carcinoma
- AUTHOR INSTRUCTIONS
- Current and emerging therapies for first line treatment of metastatic clear cell renal cell carcinoma
- Bone marrow niches in myelodysplastic syndromes
