Isolation and Identification of Pathogenic Microorganisms Causing Postharvest Spoilage in Hami Melons
ZHAO Xinxin, NING Ming, ZHANG Qin, CAI Wenchao, SHAN Chunhui, TANG Fengxian
(Food College, Shihezi University, Shihezi 832003, China)
Abstract: To investigate the relationship between microorganisms and postharvest spoilage of Hami melons, with traditional cultivation of microorganisms, we studied the dynamic changes of cultivatable microorganisms on Hami melons during storage at different temperatures. It was found that the main pathogenic bacteria causing postharvest spoilage of Hami melons were mycetes, including Fusarium, Alternaria, Penicillium, and Staphylococcus. The dominant pathogens causing spoilage in Hami melons during refrigerated transport and storage were Penicillium and Fusarium. This study indicated that postharvest spoilage in Hami melon was caused by single pathogenic bacteria and interactions among multiple pathogenic bacteria through a complicated mechanism. This study is conducive to a better understanding of the dynamics of microbial community during the storage of Hami melon fruit. Further study should be conducted to monitor characteristics of spoilagerelated species isolated from Hami melons so as to restrict the growth of pathogens and extend the shelf-life of Hami melons,which will provide theoretical foundation for improvements in the existing post-harvest preservation technology.
Keywords: Hami melon; preservation; pathogenic bacteria; spoilage
Hami melon (Cucumis melovar.saccharinus) is an elite cultivar of muskmelon that belongs to Cucurbitaceae. More than 100 varieties of Hami melon have been grown in China in the Turpan Basin and the Hami region of Eastern Xinjiang,Jiashi, Pishan, and Megeti of southern Xinjiang, as well as the five rivers and Altay in the northern frontier[1-2]. Hami melons are grown because they are an important economic crops, which are sold to the developed cities and regions in southeast China and exported to Asia, Europe and America[3].Hami melons are popular worldwide and are considered in China to be a national geographic product and the king of melons because of their sweet color, pleasant aroma, and crisp taste. However, the Hami melon is extremely vulnerable to infection and corruption by pathogenic microorganisms after harvest, which gives it a short storage period of around 60-70 days[1].
The most recent research has shown that the corruption of Hami melons originated because of infection by pathogenic microorganisms, as well as the respiration of Hami melons.Various methods, including independent or combined treatment with chitosan[4-5], chlorine, chlorine dioxide[6-7],ethylene wax[8], natamycin[9], allyl isocyanate,Streptococcus lactis, hot water, X-ray, and benzothiazide[10]have been used to prolong the shelf life of Hami melons to inhibit or delay the growth of pathogenic microorganisms[11-12]. In addition,to reduce the rot rate of Hami melons, exogenous substance can also be used to reduce the melon’s respiratory rate during storage to delay the respiratory peak time[13-15]. Therefore,it is of great significance to isolate and identify the main pathogens of Hami melon after harvest and take effective measures to reduce the storage loss and to prolong the storage period of Hami melon after harvest.
Many opportunistic and potentially pathogenic bacteria have been reported, includingStaphylococcusspp.,Clostridiumspp., and Enterobacteriaceae[16]. In particular,Salmonellaspp.,Escherichia coli, and isolates of theBacilluscereus-group have been found on the rinds of Hami melons. According to research reports, the main pathogenic microorganisms of Hami melon during postharvest storage areFusarium,Rhizopus,Cadophora,Aspergillus, andPenicillium. When Hami melon is infected byFusarium,white velveteen mycelium will emerge from the fissure,producing orange-red mucilage granules. The infection caused by variousFusariumspecies in the subdivision of Hemiptera isF. oxysporumf. sp.[17].Fusariumare widely found in air and soil. When conditions are suitable, the conidia produced can be reinfected from the mechanical wound of Hami melon. When being infected byRhizopus, the mechanical wound or fissure often has white to gray thick or sparse floss, and a little black mold appears on it, which is the fruiting body of pathogenic bacteria. Finally, water drips from the diseased part of Hami melon, which leads to rapid decay of Hami melon. Soft rot of Hami melon is caused by a variety ofRhizopusin the subgenusConjunctiva, the most important pathogen of which isR. stolonifer[18]. When Hami melon is in contact with the ground or has an injured part,Penicilliumfirst forms a dark yellow necrotic spot in the form of water stain[19], which is sunken downward. With the development of the disease, the diseased tissue gradually begins to soften and rot. The dense green mildew on the surface of the diseased spot usually only infects the over-ripe, mechanically injured or partially necrotic fruits. Of this list, the fungi belonging toFusariumandPenicilliumare most often observed. However,there has been no definitive evidence that corruption is an immediate result of a fungal infection, and the dominant pathogens and their pathogenicities remain unknown.
The deterioration of a Hami melon is always accompanied by the growth of the Hami melon, which indicates that pathogenic microorganisms are likely the main cause of the deterioration of Hami melon. Since Hami melons are vulnerable to infection and corruption by pathogenic microorganisms, technologies to maintain the freshness of Hami melons have become one of the most important challenges to developing a Hami melon industry in China.Although it has been reported that multiple microorganisms contribute to the deterioration of Hami melons[6], there is little research on the dynamic change of microbial polymorphism during the preservation process of Hami melons. In this study, we aimed to investigate the variation of microbial diversity on the skin of Hami melons at different preservation temperatures, the dynamic changes of culturable microorganisms using traditional microbiological separation technology. This study provides a theoretical basis of the improvement in the postharvest preservation technology of Hami melons.
1 Materials and Methods
1.1 Materials and reagents
1.1.1 Melon preserving
Jiashi muskmelon, a representative Hami melon from Xinjiang, was collected from Shihezi and used as the sample in this study. The harvested Jiashi muskmelon with a degree of ripeness of eight, a uniform size, and with no pest diseases or mechanical damage. According to the normal preservative method, Hami melons were washed with running tap water to remove adhering soil prior to disinfection of the surface using 70% ethanol. Then, samples were placed at (3 ± 0.5) ℃ and(21 ± 0.5) ℃ in a cold storage warehouse and were removed at 60, 67, and 74 days. The infected fraction was eluted with sterile lint and sterile water and was collected on a sterile plate.
1.1.2 Medium and reagent
Peptone, beef extract, NaCl, natamycin, agar, glucose,KH2PO4, MgSO4, C20H2Cl4I4Na2O5, chloramphenicol was purchased from Sinopharm Group Chemical Reagent Co.Ltd.; dNTPs Mix, 5 ×TransStartTM, FastPfuBuffer and FastPfuFly DNA was purchased from Polymerase Bao bioengineering Dalian Co. Ltd..
1.2 Instruments and equipment
SW-CJ-10 clean bench was purchased from Suzhou Purification Co. Ltd.; XSS-2 electron microscope was purchased from Olympus (China) Co. Ltd.; DYY-6C electrophoresis apparatus was purchased from Beijing Liuyi Biotechnology Co. Ltd.; PHS-3EpH meter was purchased from Shanghai Yidian Analytical Instrument Co. Ltd.; A300 gradient polymerase chain reaction (PCR) instrument was purchased from Hangzhou Langji Scientific Instrument Co.Ltd.; GIS-630 gel imager was purchased from Hangzhou mio instrument Co. Ltd..
1.3 Methods
1.3.1 Isolation and purification of pathogens
The pathogens were plated separately on nutrient agar(10.0 g/L peptone, 3.0 g/L beef extract, 5.0 g/L NaCl, 0.1 g/L natamycin, 15.0 g/L agar, final pH (7.3 ± 0.2)), then cultured for 1-2 d at 30 ℃, and Rose Bengal medium (5.0 g/L peptone, 10.0 g/L glucose, 1.0 g/L KH2PO4, 0.5 g/L MgSO4,0.03 g/L C20H2Cl4I4Na2O5, 0.1 g/L chloramphenicol, 20.0 g/L agar, final pH (7.2 ± 0.2)), and then cultured for 3-4 d at 28 ℃.Screening was performed by conventional culture techniques using the surface of the cantaloupe rot as a microorganism source. The culture was then streaked on an Agar plate and Rose Bengal agar plate and incubated at 28 ℃ to develop colonies for 1-2 d. The colonies that appeared were picked up independently and transferred to the medium, and the plate cultures were repeated twice. Once a pure culture was obtained, it was named according to the different conditions of the strain separation, and a total of 118 strains were obtained as pure cultures.
1.3.2 Morphological observations
Booth’s method was used to identify the morphological characteristics of the isolates[20]. The spore of each fungus was inoculated into Rose Bengal medium and cultured at 28 ℃ for 7 d, and the morphological characteristics of the colonies, microconidia, macroconidia, and sporulated cells were observed.
The macroscopic colony morphological characteristics of the 13 isolates were observed on nutrient agar after being incubated for 7 d at 30 ℃, and morphological characteristics were assigned to each isolate using Berger’s Bacterial Identification Manual.
1.3.3 Pathogenicity measurement
Three Hami melons, each with a mass of approximately 3.5 kg, were divided into one group containing 3 parallels. A bacterial suspension enriched with filter paper was incubated overnight in a shaker and then applied to the surface of the melon, and sterile water was applied to the corresponding position of each melon in the group as a control. Then the melons were covered with plastic wrap and kept at room temperature ((21 ± 0.5) ℃) and cold storage ((3 ± 0.5) ℃).After inoculation for 0, 12, 24 and 36 h, 3 parallel samples were taken from the same set of points.
1.3.4 Re-isolation of pathogens
Pathogens were placed on PDA medium. Each Petri dish contained a single isolate of one pathogen. There were six replications for each isolate from each Hami melon,which were arranged in a completely randomized design.A non-inoculated dish was used as a control. The sporulated fungi from each Hami melon were examined after incubation at 25 ℃ for 7 d.
1.3.5 DNA extraction
After the isolates were cultured on a potato dextrose agar (PDA) medium for 7 d, the mycelia of each isolate were collected in 2.0 mL centrifuge tubes and ground using a disposable plastic rod to extract genomic DNA.
Once the bacterial isolates were cultured in a nutrient agar liquid medium for 3 d, the bacterial suspension of each isolate was collected in a 2.0 mL centrifuge tube. After centrifugation, the bacterial cell pellet was collected, and genomic DNA was extracted. Finally, the extracted DNA samples were stored in a refrigerator at -20 ℃.
DNA was extracted from each specimen according to the modified urea method[21], and the extracted bacterial and eumycete DNA were sequenced by Sangon Biotech Co. Ltd.(Shanghai, China).
1.3.6 Molecular identification of pathogenic microorganisms
Bacterial DNA was extracted from the pathogenic isolates for PCR to amplify the 16S rDNA gene using the universal primers 27F (5’-AGAGTTTGATCCTGGCTCAG-3’)and 1492R (5’-GGTTACCTTGTTACGAC TT-3’), and the universal primer for eumycete DNA for amplification PCR, ITS1 (5’-TCCGTAGGTGAACCTGCGG-3’) and ITS4 (5’-TCCTCCGCTTATTGATAT GC-3’). The PCR reaction volume was 25 μL, including 12.5 μL of 2×mix enzyme, 0.5 μL of primer, 1.5 μL of DNA template, and 10 μL of double-distilled water (ddH2O). The eumycete DNA PCR reaction procedure was performed as follows:pre-denaturation was performed at 94 ℃ for 3 min, followed by 35 cycles of denaturation at 95 ℃ for 30 s, annealing at 55 ℃ for 30 s, an extension at 72 ℃ for 30 s, and finally an extension at 72 ℃ for 6 min. The procedure for the bacterial DNA PCR reaction was as follows: pre-denaturation was performed at 95 ℃ for 5 min, followed by 35 cycles of denaturation at 94 ℃ for 45 s, annealing at 55 ℃ for 45 s, and extension at 72 ℃ for 1 min 30 s, and finally an extension at 72 ℃ for 5 min 30 s. The PCR products were separated and purified by performing gel electrophoresis with 1.0% agarose.Of the 107 isolates confirmed as non-repetitive, 69 were identified at the species level.
1.3.7 Phylogenetic analysis
MEGA 7.0.21 software was used during the phylogenetic analysis based on 16S rDNA or ITS sequences of the isolated strains. Homogeneous sequences of strains were obtained from the GeneBank database and were then analyzed on the NCBI standard nucleotide BLAST. Sequence analysis enabled the isolates to be identified at the species level based on their percentage identity. The pathogens were then identified based on their similarity to records in the GenBank database by using the online BLAST tool.Neighbor-Joining (NJ) methods of MEGA software were used to construct phylogenetic trees according to sequence construction methods. The bootstraps of all branches were tested in over 1 000 replicates, and the phylogenetic tree was classified by genus[22].
1.4 Statistical analysis
The incidences of spoilage were calculated as the mean of the replicates (n= 3). Differences between the 107 isolates within poilage were calculated using the statistical software package SPSS 13.0. Analysis of variance (ANOVA) with Duncan’s multiple range test and Dunnett’s tests method atP< 0.01 were performed. A general linear model was used to perform an interaction analysis of spoilage and isolates.
2 Results and Analysis
2.1 Isolation and selection of pathogenic microorganisms causing post-harvest spoilage in Hami melons
A total of 118 isolates (74 fungi and 44 bacteria) were collected from 60 samples taken from the 121 Mission Farm in Shihezi, Xinjiang over period of 74 d. The results from the BLASTn alignment of the individual isolates identified 14 different genera, comprised of multifarious species within each genus. When comparing the internal transcribed spacer(ITS) rRNA sequences from isolates obtained in our research to the BLASTn, 45 isolates showed > 98% similarity.
Pure cultures from diseased Hami melons were isolated on Rose Bengal medium and were numbered CCaDW.4,CCaDW.5, CCaDW.9, CCaDO.1, CDW.1, CDW.3, CUY, and CUG+B.3, respectively. Each culture had similar features, and after being cultured for 7 d, the colonies were 40 to 46 mm in diameter.Their hyphae were floccose, and white to pinkish at the back sides (Fig. 1). They produced many microeonidias which was round or oval, and a small number of macroconidia which were sickle-or spindle-shaped, with slightly curved terminal cells. Initially, these fungal isolates were considered to beFusariumspp., according to their morphological characteristics. They were labelled 3 ℃-CaUA.1, 3 ℃-U.2,3 ℃-CaDA.1, 3 ℃-CaDA.2, 3 ℃-CaDAQ, CCaDW.2,3 ℃-DG.2, CCaD.5, 3 ℃-CaD.4, CCaDO.2, 3 ℃-CaUA.2,3 ℃-DJ.2, 3 ℃-CaU.4, CaDAQ.6, and CCaDG. The colony colors varied from light to dark greenish gray, including grayish, pale, pale yellowish and olive green, whereas the colony textures varied from velvety to fasciculate, including weakly floccose. There was a considerable variation in their size, and some isolates exhibited a very thin margin,whereas others displayed a thick margin which comprised up to one third of the colony’s diameter. These fungal isolates were identified asPenicilliumspp., and theAspergilluswas morphologically identified based on the black or yellow colony. The isolates numbered 3 ℃-CaD.1, 3 ℃-CaU.5,3 ℃-DJ.1, and CUG+B were separated into two groups since the appearance of the colonies were slightly different from one another. Two isolates produced compact black colonies with a greyish central region and occasionally produced slightly yellowish colonies and a brighter reverse colony appearance, whereas two others grew hugely on the plate with uniform black to greyish black colonies. The surface of the colony was gray short-fleece aerial hyphae, which were raised in the center with a circle of immersed hyphae at the edge. The isolates numbered CCaDQ.1, CCaDQ.2, CCaDQ.5,CCaDQ.6, CCaDW.1, CCaDW.3, CCaDW.6, CCaDW.10,CCaDY, and CCaDA.1 were identified asCladosporiumspp..Meanwhile, the mycelium of the genusMucordeveloped and branched into a spider web and was white with a black sporangium. According to its morphological characteristics,the isolates numbered CCaDG.3, CCaD.1, CCaD.2, CCaD.8,CCaDW.11, 3 ℃-CaD.5, and CCaDW.5 were identified asMucorspp.. Additionally, according to the Berger’s Bacterial Identification Manual, the bacterial isolates were identified asStaphylococcusspp.,Enterococcusspp.,Stenotrophomouasspp.,Curtobacteriumspp.,Bacillusspp.,Exiquobacteriumspp.,Pantoeaspp.,Acinetobacterspp., andSteuotrophomonasspp. (Fig. 2).



Fig. 1 Variations in fungal community composition at different stages of storage
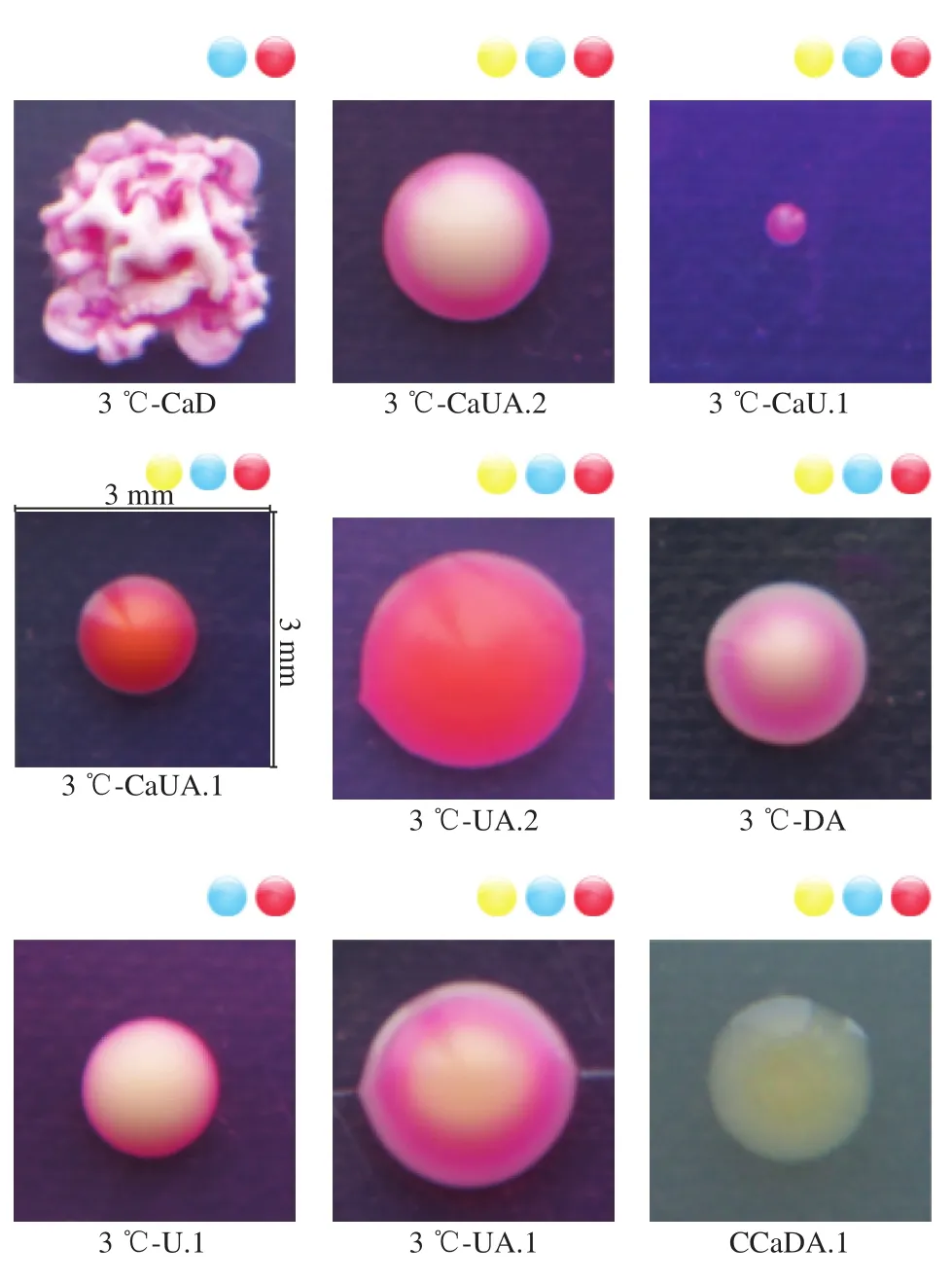
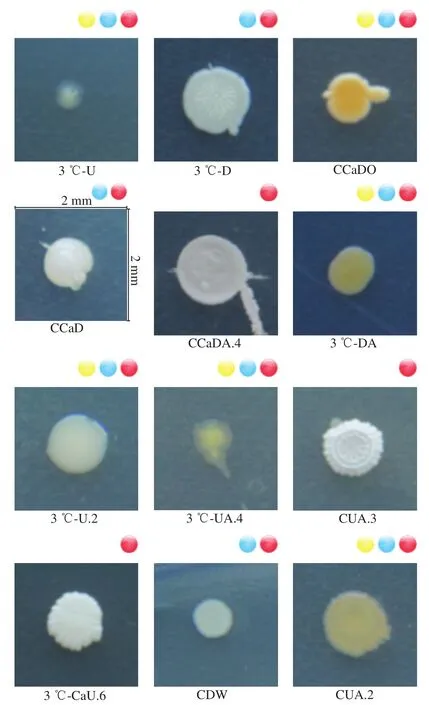
Fig. 2 Changes in the microbial communities of Hami melons during storage
2.2 Molecular identification
Phylogenetic analysis of the 16S rRNA sequence data resembled the BLASTn results, which formed 8 distinct clusters includingStaohylococcus,Enterococcus,Stenotrophomouas,Curtobacterium,Bacillus,Exiquobacterium,Pantoea,Acinetobacter,andSteuotrophomonas. Closely-related genera, such asPantoeaandEnterococcusspecies, were not easily differentiated within the 16S rRNA phylogenetic tree and were simply grouped together instead of being defined in groups. Of the 21 isolates sequenced, isolates appeared to belong to the genusStaohylococcus(13, 20, 23, 27, 112, 114, 115 and 119),Pantoea(10, 14 and 29),Curtobacterium(1 and 121),Bacillus(3 and 7),Enterococcus(15 and 24), while the additional four isolates were grouped in the known genus withAcinetobacter,Steuotrophomonas,ExiquobacteriumandStenotrophomouas. In addition, a search of sequences in the NCBI database revealed 3 isolates that were the most homologous toB. amyloliquefaciens(Fig. 3a), with a homology greater than 99%. Isolates 13, 20, 28, 29, 112, 114,115 and 119 were 100% homologous toS. succinus(Fig. 3b),Exiguobacterium indicum(Fig. 3c),P. dispersa(Fig. 3d).

Fig. 3 Phylogenetic trees of strains 3 (a), 13-20-112-114-115-119 (b),28 (c) and 29 (d)
Sequencing of the ITS rRNA region was also performed for 24Penicilliumisolates, 11Fusariumisolates, 4Asoergillusisolates, 4Cladospriumisolates, 2Mucorisolates,and 2Gibellulopsisisolates. The PCR amplified 18S rRNA genes from the isolated fungus were sequenced and analyzed using the BLAST program of the NCBI. The BLAST results showed that the 18S rRNA gene sequences of isolates exhibited 98% to 100% similarity to known sequences.

Fig. 4 Phylogenetic trees of strain 52 (a), 61-71 (b),78-79-96-98 (c) and 105 (d)
During phylogenetic analyses based on 18S rRNA gene sequences, twenty-four isolates showed a strong similarity toPenicillium; eleven isolates were similar toFusarium; four showed incredible similarity inCladosporium; four isolates were similar toAspergillus; two isolates were similar toMucor; and the additional two isolates were grouped with the known genus ofGibellulopsis. The 18S rRNA gene sequence from isolate 52 showed 99% sequence similarity withF. pseudoanthophilum(Fig. 4a), while isolates 61-71 showed 98% similarity withM. nanus(Fig. 4b), isolates 78-79, 98 showed 100% similarity withP. crustosum(Fig. 4c),and isolate 96 showed 100% similarity withP. polonicum(Fig. 4c), while isolate 105 showed >99.9% sequence identity toC. endophytica(Fig. 4d).
2.3 Pathogenicity test results
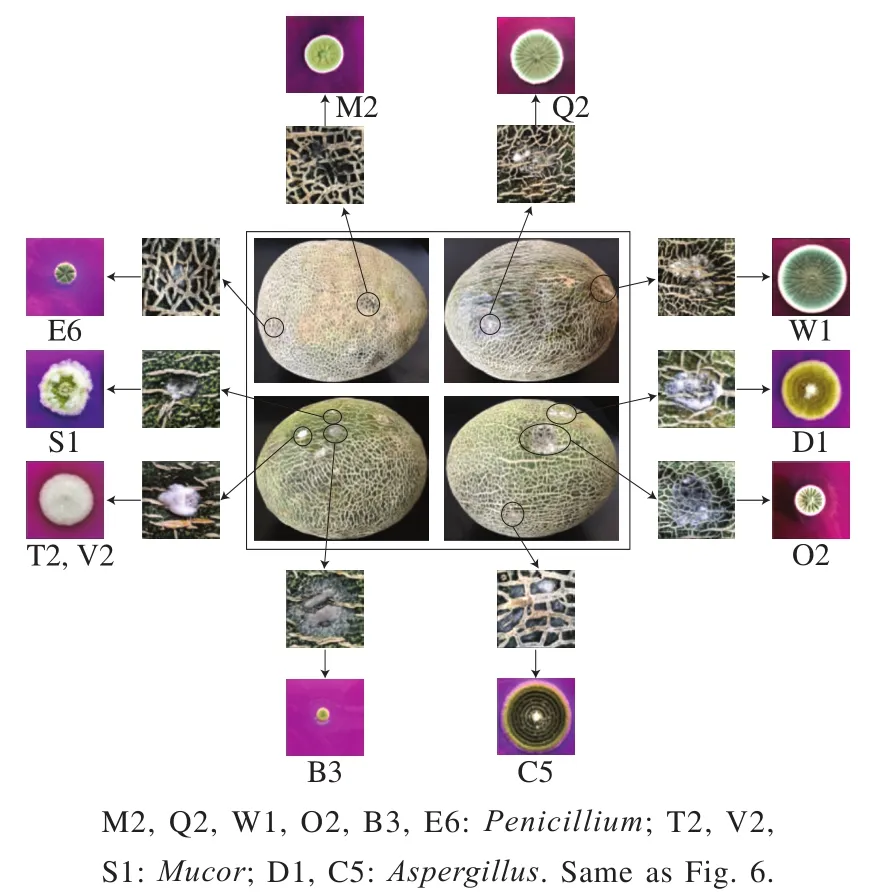
Fig. 5 Dominant microfloras during the pre-vaccination period
Reverse inoculations were performed as a quick and reliable method to select pathogenic bacteria for additional screening of Hami melons.Penicillium,Mucor, andAspergilluswere the dominant flora present during the preinoculation period (Fig. 5). Replicated inoculations were similar across all experiments and are presented in Fig. 6.Penicillium,Mucor, andFusariumsubstantially reduced the quality of the Hami melon and putridity symptoms began to be displayed 3 d after inoculation, whereas the melon quality was not affected by the strainsGibellulopsisandCladosporium. After the Hami melons were inoculated,pathogenic bacteria were re-isolated from symptomatic tissue,which was consistent with previously-isolated pathogens, thus fulfilling Koch’s postulates. During the same experiment,the quality of Hami melons was reduced byPenicilliumtwo days after the initial inoculation. Melon quality was not seriously reduced by any other species and isolates tested when compared to the non-inoculated control. Three days after inoculation, isolates ofMucorandFusariumwere shown to reduce melon quality. At the same time, the Hami melon quality of plants inoculated withGibellulopsisandCladosporiumshowed no differences from the non-inoculated control. Reductions in Hami melon quality were observed as sunken areas on the melon surface and undermining.

Fig. 6 Results of pathogenicity test
3 Discussion
According to the research reports of Ozbahce[23]and Suzuki et al.[24], Hami melon can be latently infected by pathogenic bacteria during the whole growth and development process, of which reticulate period is an important period of pathogenic bacteria infection. Gautam et al.[25]investigated the interactions betweenSalmonella entericaPoona, the melon pathogenic bacteriumErwinia tracheiphila, and cantaloupe fruit. The study was conducted by inoculating fruit surfaces with pathogenic bacteria during the natural cracking stage either independently or together, over a 2 cm × 2 cm area of rind that contained a crack. Liu Tao et al.[26]determined that the main pathogenic fungi causing postharvest decay of Hami melon in Xinjiang includeFusarium,Stachybotrys,PenicilliumandGibberella,which are consistent with the results of this study. Chen Cunkun et al.[27]have isolated the pathogenic bacteria with strong infectivity from the main Hami melon varieties in Xinjiang through research. The results show that the main pathogenic bacteria causing decay and deterioration of Hami melon during post-harvest storage includePenicillium,Cladosporium,Aspergillusand so on. Previous studies have demonstrated that the main spoilage microbial species on a variety of Hami melons wasPenicillium. In the present work,we observed thatPenicilliumdominated the microbiota after a storage period of 67 d. The rapid growth ofPenicilliumcould be due to a higher competitivity compared to other pathogenic bacteria under the current storage conditions(3 ℃). Either a strain or a microbial group was collectively responsible for melon spoilage, but not all species or strains are necessarily responsible for the spoilage. Generally,spoilage is attributed to the dominant microbiota, which are usually referred to as specific spoilage organisms (SSO)[28-29].Penicilliumwas frequently discovered to be the dominant reference species, and would likely be considered the SSOs,which was recognized since SSOs may be associated with the spoilage of Hami melons during storage. However, the putridity mechanisms and characteristics should be investigated further.Additionally, the multiformity showed thatCladosporiumsteadily increased after day 67, and significant changes alternately occurred in the predominant microbiota ofCladosporiumandPenicilliumon day 74, whenCladosporiumunderwent a dramatic increase and then became the dominant bacteria (21 ℃). Furthermore,Aspergilluswas isolated on the 60thday and then increased on day 67 and afterwards reached its stationary phase and then declined but still remained at a relatively high level. Previous studies have shown thatAspergillusis frequently isolated from a variety of foods and are frequently the predominant members of microbial communities found in foods[29]. Previous research has also shown thatAspergillusalso have been detected in melons, and although theAspergillusare frequently isolated from melons,they are rarely present in large numbers. The results presented in the current study agree with these results.Aspergillusis considered to possess extensive potential for melon spoilage and is important players in Hami melon storage. In addition to mold, Hami melon spoilage is often associated with bacteria such asStaohylococcus,Enterococcus,Stenotrophomouas,Curtobacterium[30],Bacillus[31],Exiquobacterium,Pantoea[32],andAcinetobacterandSteuotrophomonas. In the present study,Staohylococcuswas present in abundance at all points of storage, but it is not an important member of the microbial community that is typically associated with spoilage[33].
Hami melon can be latently infected by pathogenic bacteria during the whole growth and development process,which eventually leads to the decay and deterioration of the harvested Hami melon and makes the Hami melon lose its nutritional quality and commercial value. Moreover, there has been a report that the microbial populations of molds and yeasts were significantly higher on the lower surface of the Hami melon that is in direct contact with the soil[34].This research has shown that an uneven distribution of microbial populations exists on the surfaces of field-grown Hami melons, which suggests that direct contact with soil is likely a major source of microbial melon contamination.Field production practices should be undertaken to minimize direct contact of Hami melon with the soil to ensure a safer product for the consumer, and more effective surface cleaning methods should also be explored.
As early as the early 1960s, some scholars proposed that pathogenic bacteria could produce cell-wall degrading enzymes (cell-wall degrading enzymes, CWDE). Cellwall degrading enzymes play a very important role in the pathogenic process of plant pathogenic bacteria. Studies have found that the activities of various cell-wall degrading enzymes in some diseased plants infected by pathogenic bacteria are much higher than those in healthy tissues.During the development of the same disease, various cell wall degrading enzymes appear in different time modes and play different roles. Plant cell tissues primary wall and secondary wall contain more polysaccharides, which can be degraded by cell wall degrading enzymes produced by pathogenic bacteria. The pathogenic bacteria invade plant cells and spread in host tissues, which is the main cause of plant diseases. In the process of pathogenic bacteria infecting plant tissues, their pathogenic factors can sometimes act on plant tissues alone, and sometimes require synergistic effects among various mechanisms. Isoenzymes are the enzymes that first interact with each other, and the isoenzymes of pectate lyase have mutually reinforcing effects. Pectin esterase and galactopyraldehyde have synergistic effects,and the two enzymes produced by pathogenic bacteria also have synergistic effects. Pathogenic bacteria can secrete many different kinds of cell wall degrading enzymes in the process of invading plant tissues. The cell wall components of plants are components of plant tissues, which are the main barriers to prevent pathogenic bacteria from invading host tissues. The cell wall components of the cell wall degrading enzyme can destroy host plants. A series of experimental results have shown that the cell wall degrading enzymes have a very significant pathogenic effect on plant tissues.Cell wall degrading enzymes play a very important role in the pathogenic process of plants. Cell wall degrading enzymes secreted by plant pathogenic fungi can cause plant tissue infection and damage. The cell wall degrading enzyme produced by plant pathogenic fungi in the process of infecting plant tissues will lead to the destruction of plant tissues. The destruction mechanism to host tissues has been discovered, which makes the important role of cell wall degrading enzyme in the pathogenic process of pathogenic bacteria to plant tissues proved by some domestic researchers through the use of electron microscope technology. However,for a specific disease, the specific mechanism of action on plant tissues during its onset is not very clear. As far as a particular plant disease is concerned, its mechanism of action in the specific pathogenesis of plant tissue is not particularly clear. Exploring the interaction between pathogenic bacteria and plant tissues and studying the pathogenic mechanism of pathogenic factors of pathogenic bacteria have always been an important part of researchers’ efforts. Up to now, although there have been reports on pathogenic bacteria causing other fruits or plants to rot, there are relatively few reports on the specific pathogenesis of cantaloupe rot. With the continuous progress and development of social science and technology,the specific action mechanism of cell wall degrading enzyme should be clear, which not only lays the foundation for studying the molecular mechanism of pathogenic bacteria,but also has great significance for establishing new disease control measures.
4 Conclusion
In this study, 74 strains of fungi, which belong toFusarium,Penicillium,CladosporiumandMucor, were preliminarily identified by isolating and purifying pathogenic microorganisms causing the deterioration of Hami melon epidermis during postharvest storage, combining with disease characteristics and microscopic observation. Additionally,according to the Berger’s Bacterial Identification Manual,the bacterial isolates were identified asStaphylococcus,Enterococcus,Stenotrophomouas,Curtobacterium,Bacillus,Exiquobacterium,Pantoea,Acinetobacter, andSteuotrophomonas. In the reverse inoculation verification experiment,Penicillium,Mucor, andFusariumsubstantially reduced the quality of the Hami melon and putridity symptoms began to be displayed 3 d after inoculation,whereas the melon quality was not affected by theGibellulopsisandCladosporiumstrains.

