What do we know about the role of lncRNAs in multiple sclerosis?
Viviana Nociti , Massimo Santoro
Abstract Multiple sclerosis is a chronic, inflammatory and degenerative disease of the central nervous system of unknown aetiology although well-defined evidence supports an autoimmune pathogenesis. So far, the exact mechanisms leading to autoimmune diseases are still only partially understood. We know that genetic, epigenetic, molecular, and cellular factors resulting in pathogenic inflammatory responses are certainly involved. Long non-coding RNAs (lncRNAs) are non-protein coding transcripts longer than 200 nucleotides that play an important role in both innate and acquired immunity, so there is great interest in lncRNAs involved in autoimmune diseases. The research on multiple sclerosis has been enriched with many studies on the molecular role of lncRNAs in the pathogenesis of the disease and their potential application as diagnostic and prognostic biomarkers.In particular, many multiple sclerosis fields of research are based on the identification of lncRNAs as possible biomarkers able to predict the onset of the disease, its activity degree,its progression phase and the response to disease-modifying drugs. Last but not least,studies on lncRNAs can provide a new molecular target for new therapies, missing, so far, a cure for multiple sclerosis. While our knowledge on the role of lncRNA in multiple sclerosis has recently improved, further studies are required to better understand the specific role of lncRNAs in this neurological disease. In this review, we present the most recent studies on molecular characterization of lncRNAs in multiple sclerosis disorder discussing their clinical relevance as biomarkers for diagnosis and treatments.
Key Words: antisense lncRNAs; enhancer lncRNAs; epigenetics; immune system;intergenic lncRNA; intronic lncRNA; multiple sclerosis; sense lncRNAs; single nucleotide polymorphisms
Introduction
Multiple sclerosis (MS), the second leading cause of sustained neurological disability in young people after trauma (Ontaneda et al., 2017), is a chronic, inflammatory and degenerative disease of the central nervous system (CNS) (Naegele et al.,2014) of unknown aetiology. So far, there are two theories on its pathogenesis: the “outside-in” autoimmune hypothesis,for which MS is an exclusive autoimmune inflammatory disease caused by unregulated auto-reactive immune cells that from the periphery go into the CNS parenchyma,attacking various cell types, and the “inside-out” hypothesis,for which MS is a primary degenerative disease in which inflammation is secondary to a release of auto-antigens promoting autoimmunity (Stys and Tsutsui, 2019). Until now, it remains to be determined whether inflammation is primary or secondary to a degenerative process in the brain.Nevertheless, well-defined evidence (as well as the successful use of immunomodulatory drugs in reducing clinical relapses and/or neuroradiological ‘activity’) demonstrates that an uncontrolled inflammatory response in the CNS is destructive in MS (Thompson et al., 2018). Sustained disability, however,is due to a progressive neurodegenerative process, causing axonal loss and brain atrophy, primary or secondary to the peripheral and compartmentalized inflammation in the CNS(Machado-Santos et al., 2018). To date, no approved therapy has provided marked neuroprotective effects nor antiinflammatory therapies, used in the treatment of the disease,showed efficacy in the progressive phase of MS.
Well-defined evidence showed that the MS pathophysiology is characterized by altered bidirectional interactions among several immune cell types in the periphery and resident cells of the CNS, such as microglia and astrocytes (Li et al., 2018).The MS relapses, occurring in the early phases of the disease,are characterized by the infiltration, into the CNS parenchyma,of pro inflammatory CNS-specific effector T, B and myeloid cells, which are activated and/or regulated in an aberrant way(Dendrou et al., 2015). The altered function of regulatory T(Treg) cells and resistance of CNS-specific effector T cells to Treg cell-mediated regulation could be one possible cause of the neuro-inflammation (Kaskow et al., 2018; Kitz et al.,2018). Furthermore, CNS-resident cells, that secrete many inflammatory mediators, recruit inflammatory cells into the CNS (Dendrou et al., 2015). Therefore, both peripheral and CNS-compartmentalized inflammatory mechanisms contribute to MS pathogenesis (Filippi et al., 2018). In the advanced stages of the disease, infiltrated immune cells into the CNS are few and ongoing CNS-compartmentalized inflammation seems to dominate progressive phases of MS. During this phase, the role of B cells in driving inflammation seems to be prominent,particularly within meningeal inflammation (Magliozzi et al.,2007; Correale et al., 2017).
Until now, the exact mechanisms leading to autoimmune diseases are still only partially understood but we know that genetic, epigenetic, molecular, and cellular factors resulting in pathogenic inflammatory responses driven by self antigenspecific T-cells are certainly involved.
Whole-genome transcriptional analysis have recently shown that the genome, in eukaryotic cells, can be transcribed in numerous types of coding and non-coding RNAs, the letter constituting at least 90% of these RNAs (Djebali et al.,2012). Increasing evidence on non-coding RNAs (ncRNAs)demonstrated that, more than evolutionary “junk genes”,they have an important role as regulators of different cellular processes also in many diseases (Sun et al., 2013). ncRNAs are grouped into small ncRNAs (< 200 nucleotides) and long ncRNAs (≥ 200 nucleotides).
Long-ncRNAs (lnc-RNA) have more extensive and complex regulatory mechanisms than small ncRNA (Schmitz et al.,2016). Among their several functions (cell proliferation and differentiation, immune responses, metabolism, and apoptosis), they are very important in human autoimmune diseases playing specialized roles in modulating immune cell differentiation and activation (Sigdel et al., 2015; Wu et al.,2015; Atianand et al., 2017).
Indeed, long non-coding RNAs (lncRNAs) have an important impact on both innate and acquired immunity (Sigdel et al.,2015; Atianand et al., 2017; Zhang et al., 2017).
Innate immune responses are the body’s non-specific first line defence against pathogenic microorganisms, by the action of macrophages, dendritic cells, and natural killer cells.LncRNAs may have a critical role in regulating this response to pathogens (Mao et al., 2015; Atianand et al., 2017; Ivanov et al., 2018). Many changes have been found in lncRNA expression in macrophages upon innate immunity activation(Hu et al., 2016; Tong et al., 2016; Yang et al., 2016; Fei et al., 2017; Ye et al., 2018) and in dendritic cells are antigenpresenting cells (Ahmad et al., 2020). LncRNAs also have an important role in regulating the acquired immune responses, a second line of defence against pathogens, producing antigenspecific responses and immunological “memory.” Lymphocytes T and B are the main immune cells of the adaptive immune system. T helper cells are also critical in the pathogenesis of several diseases, and overall in autoimmune diseases (Zhu et al., 2010). Important lncRNA expression patterns have been elucidated in T cell function (Atianand MK et al., 2017) and in distinct stages of B cell development (Petri et al., 2015; Brazao et al., 2016; Tayari et al., 2016). LncRNAs also have a key role in cytokine genes regulation but overall the antisense lncRNAs(Atianand et al., 2017).
Currently, many MS fields of research are based on the identification of possible biomarkers able to predict the onset of the disease, its activity degree, its progression phase and the response to disease-modifying drugs. Among the various biomarkers analyzed in MS, lncRNAs are of great importance because they are able to regulate the genome expression/stability and cellular functions such as proliferation,differentiation, apoptosis and development especially in the activation of immune cells both in innate and in adaptive immune system (Esteller, 2011; Wapinski and Chang, 2011).Indeed, it has been demonstrated that unregulation of lncRNAs may have a crucial role in the pathogenesis of autoimmune disorders such as systemic lupus erythematosus,rheumatoid arthritis and psoriasis (Sigdel et al., 2015).
In this review, we present the most recent studies on molecular characterization of lncRNAs in the MS disorder discussing their clinical relevance as biomarkers for diagnosis and treatments.
Search Strategy and Selection Criteria
We have performed a PubMed literature search of articles with search terms including “multiple sclerosis”,“pathogenesis”, “immune system”, “autoimmune diseases”,“epigenetic”, “non-coding RNA”, “long non-codingRNA”, “single nucleotide polymorphisms”.
Selection criteria included recent articles (2011-2020) on MS pathogenesis, role of lncRNA in autoimmune diseases and recent findings on lncRNAs in MS.
Biogenesis and Function of Long Non-coding RNAs
LncRNAs are a group of non-coding RNA (ncRNAs) transcribed by RNA polymerase II and RNA polymerase III at several loci of the genome (Beermann et al., 2016).
Generally lncRNAs are more than 200 nucleotides in length and exhibit mRNA-like features such as splicing,polyadenylation (poly-A+) tail and 5′ capping (5′cap) but without protein coding capacity (Derrien et al., 2012).
However, recent studies identified same lncRNA with an open reading frame (ORF) and few exons suggesting with a high probability translational ability to encode proteins (Bazin et al., 2017; Wang et al., 2017).
LncRNAs showed a tissue and cell specific expression, so that their levels are influenced by developmental or physiologic and pathologic state (Darren et al., 2012; Ulitsky and Bartel,2013).
Comparing with mRNA, lncRNAs presented a lower expression in all tissues so that they were initially considered transcriptionalnoiseresulting from low RNA polymerase fidelity (Darren et al., 2012). With the advent of highthroughput sequencing such as RNA-sequencing (RNA-seq),several studies showed that lncRNAs are abundant in the human genome and they can regulate the gene expression by DNA, RNA and protein interaction affecting transcriptional,post-transcriptional and translational processes (Atianand and Fitzgerald, 2014).
LncRNAs biogenesis occurs in both the nucleus or the cytoplasm and their cellular localization can influence the regulation mechanism of lncRNAs (Figure 1).
In the nucleus the lncRNAs are mainly involved in transcriptional regulation, through epigenetic modulation including histone and chromatin structure alteration that limits DNA accessibility(Tang et al., 2017;Figure 1). Instead, in the cytoplasm the lncRNAs are mainly involved in post-transcriptional regulation and post-translational regulation through mechanisms of translation inhibition, alternative splicing regulation and degradation (Tang et al., 2017;Figure 1).
LncRNAs classification is based according to their genomic position including proximity to protein-coding genes although several lncRNAs do not fall into this classification but rather turn out to be a combination of these specific characteristics or they cover long genomic sequences (Kung et al., 2013).
The main lncRNA classes are:
1) Intergenic lncRNAs (lincRNAs) represent the largest and most representative group of lncRNAs located among protein coding genes (Xue et al., 2017). By RNA-seq several lincRNAs have been identified in the genome showing a low level of expression (Cabili et al., 2011). They present few exons and can regulate gene transcription in a cell-specific manner by acting either incisor intransregulation (Cabili et al., 2011)(Figure 1).
2) Intronic lncRNA (ilncRNA) are entirely derived from the introns of transcript gene in either sense or anti-sense direction. These lncRNAs have been associated with the miRNAs, small nucleolar RNAs (snoRNAs) and circular noncoding RNAs (circRNAs) (Zhang et al., 2013).

Figure 1|The biogenesis of lncRNAs involves both the nucleus and the cytoplasm with specific transcriptional and post-transcriptional functions.The lncRNA classification is based on their transcriptional origin: intergenic lncRNA(lincRNA) transcribed from the genomic region between two genes; intronic lncRNA(ilncRNA) transcribed from the introns;enhancer lncRNAs (eRNAs) transcribed from genomic regions that contain regulative sequences of the gene; sense lncRNAs transcribed from the sense strand of protein-coding gene overlapping partially or completely with intron and exons; anti-sense lncRNAs or natural antisense transcripts(NATs) transcribed from the exons of the protein coding gene on the opposite strand with partial to complete overlapping.lncRNAs: Long non-coding RNAs.
The ilncRNA functions and their regulation mechanisms are unclear and relatively unexplored. However, these lncRNAs are transcribed together with their host coding gene, thus they could probably regulate the expression of the host gene(Boivin et al., 2018;Figure 1).
3) Enhancer lncRNAs (eRNAs) includes promoter-associated lncRNAs, untranslated region-associated lncRNAs and telomere-associated lncRNAs. Indeed, they are transcripted bi-directionally in both polyadenylated or non-polyadenylated forms from active enhancer genomic regions (Xu et al.,2019). These regions are located regulative sequences of the gene contributing to the transcription start by the binding of transcriptional factors.
In this way, eRNAs can modulate the promoter activity,enhancer interactions and chromatin conformation influencing the transcription of neighboring genes (Chen et al., 2017; Liu et al., 2017;Figure 1).
4) Sense lncRNAs are transcribed from the sense strand of protein-coding gene and their sequence overlap, partially or completely, with the entire sequence of a protein coding gene including intron and part of exons (Devaux et al., 2015;Figure 1).5) Anti-sense lncRNAs or natural antisense transcripts are encoded from exons of protein-coding genes in the opposite strand with partial to complete overlapping (Devaux et al.,2015). Anti-sense lncRNAs can modulate neighbouring genes expression through a regulation incis(Magistri et al., 2012).About 70% of coding genes have anti-sense counterparts(Villegas et al., 2015;Figure 1).
The lncRNAs have the ability to inhibit or promote the expression of coding genes in tissue or cell-type manner and stage-specific development, suggesting their involvement in the pathogenesis of several diseases such as autoimmune and neurological disorders (Ingwersen et al., 2015; Sigdel et al.,2015).
In the context of autoimmune diseases, the research on MS has been enriched with many studies on the molecular role of lncRNAs in pathogenesis of this disorder and their potential application in MS such as diagnostic and prognostic biomarkers (Yang et al., 2018; Li et al., 2020).
Expression Profile of Long Non-coding RNAs in Multiple Sclerosis
In the last years, many studies have provided evidence of how lncRNAs deregulation is involved in the pathogenesis of MS disease as shown inTable 1. Here, we show the most recent studies on lncRNAs expression in the MS disease considering the biological material [serum, plasma, peripheral blood mononuclear cells (PBMC) and blood] on which the analysis was conducted. Serum represents one of the most accessible biological samples and provides excellent materials for the study of possible disease biomarkers. Recently, Santoro et al.,screened 84 lncRNAs, involved in autoimmunity and human inflammatory response, in serum from 8 healthy controls, 16 secondary progressive MS (SPMS), 12 primary progressive MS (PPMS) Italian patients (Santoro et al., 2020). The authors found the up-regulation of taurine up-regulated 1 (TUG1)in SPMS patients, while PPMS patients showed a downregulation of non-protein coding RNA 188 (LRRC75A-AS1)and a significant up-regulation of long intergenic non-protein coding RNA 293 (LINC00293) andRP11-29G8.3(Santoro et al.,2020).In-silicoanalysis with bio-informatics tools to predict the interaction of lncRNAs-miRNAs such as DIANA-LncBase v2 and HMDD v3.0 software (Paraskevopoulou et la., 2016;Huang et la., 2019), identified 21 miRNAs prediction targets of these four lncRNAs that are involved in MS disease (Santoro et al., 2020).
In previous papers, the same authors showed the specific over-expression of three circulating lncRNAs in the serum of 12 relapsing-remitting MS (RRMS) patients: nuclear paraspeckle assembly transcript 1 (NEAT1), 7SK small nuclear RNA (RN7SK RNA) andTUG1(Santoro et al., 2016). These three lncRNAs are involved in specific regulatory functions:NEAT1promotes the increase ofCXCL8expression of the gene encoding interleukin 8 via relocation of SFPQ splicing factor(Imamura et al., 2014),RN7SK RNAis involved in regulation of CD4+T lymphocytes (Sung et al., 2006) andTUG1is a component of the p53 regulatory network (Rossi et al., 2014).
Although both represent pilot studies given the limited number of samples and need confirmation in larger collection of RRMS, SPMS and PPMS samples, data obtained suggest the potential role of these lncRNAs in progressive MS pathogenesis. Another recent study conducted on serum of 60 healthy controls, 42 RRMS and 18 SPMS Egyptian patients showed an up-regulation ofLincR-Gng2-5′ ASand a downregulation ofLincR-Eрas1-3′AS, two anti-sense intergenic lncRNAs located in T helper 1 (TH1) and in T helper 2 (TH2)cells (Shaker et al., 2020). Furthermore, the unregulation of these two lncRNAs was more marked in SPMS than in RRMS patients with an opposite correlation regarding the Expanded Disability Status Scale (positive forLincR-Gng2-5′ASand negative forLincR-Eрas1-3′AS) (Shaker et al., 2020).
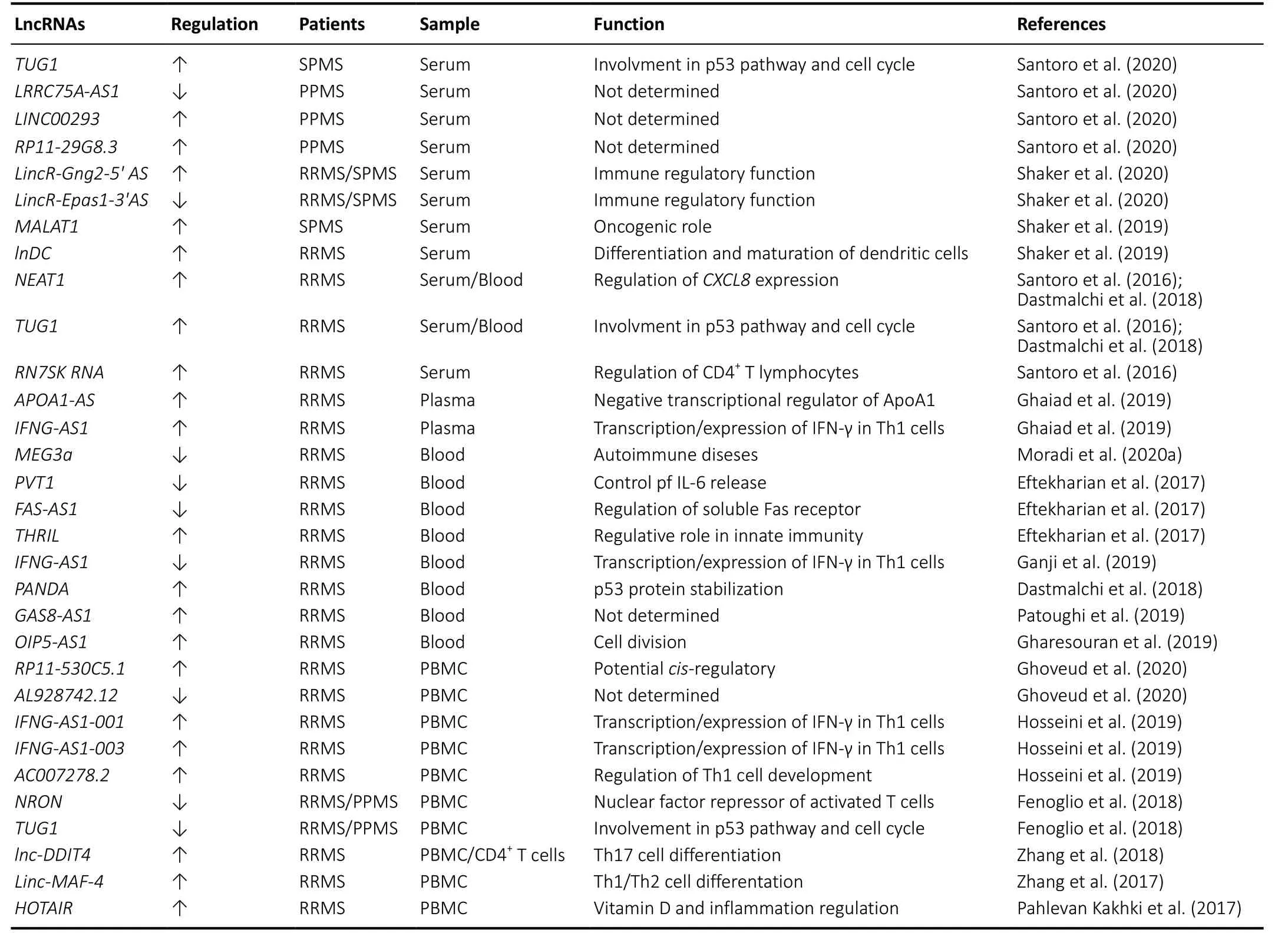
Table 1 |LncRNAs dysregulated in multiple sclerosis
Moreover, in a previous study, Shaker et al. (2019) analyzed the expression levels of metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) and lnDC lncRNAs in serum from 45 MS patients and 45 controls finding an up-regulation ofMALAT1in the SPMS subgroup with no significant differences in RRMS patients. Indeed,lnc-DCshowed an opposed expression: up-regulated in RRMS patients and no changes in SPMS subgroup (Shaker et al.,2019). In both studies, the expression analysis was performed only on some specific lncRNAs making it impossible to know if other lncRNAs are also involved.
Regarding the lncRNAs expression analysis in plasma, we found only one paper by Ghaiad et al., (2019) that determined the expression levels of anti-sense (AS) lncRNAsAPOA1-ASandIFNG-AS1in the plasma of 72 RRMS Egyptian patients(37 during relapse and 35 in remission) and 28 healthy controls. The authors found a significant up-regulation ofAPOA1-ASduring relapse and in remission comparing with healthy controls while ApoA1 and high-density lipoproteinscholesterol levels were significantly lower in the same phase,together with higher low-density lipoprotein-cholesterol levels(Ghaiad et al., 2019).
Moreover, the expression levels ofIFNG-AS1were also significantly higher in RRMS patients during relapse and in remission comparing with healthy controls (Ghaiad et al.,2019).
This study provides evidence for diagnostic and prognostic role ofAPOA1-ASandIFNG-AS1in RRMS patients although further analysis is needed for validation in a larger cohort of MS patients.
Whole blood is also often used to evaluate lncRNA expression levels. Moradi et al. (2020a) analyzed in the blood of 10 controls and 20 Iranian RRMS patients, the expression profile ofMEG3a,AC000061.1_201, andAC007182.6, three lncRNAs involvement in the pathogenesis of human autoimmune diseases. The authors found a down-regulation only ofMEG3athat negatively correlate with Expanded Disability Status Scale.Additionally, analysis of receiver operating characteristic curve showed the ability of this lncRNA to discriminate between RRMS and healthy individuals suggestingMEG3a(Moradi et al., 2020a). Even ifMEG3ais indicated as a potential diagnostic biomarker to distinguish MS patients, this study was conducted on a small cohort of samples (10 controlsvs. 20 RRMS patients) and further experiments are needed to validate these results in a large number of individuals considering also the different stages of the MS disease.
Another blood study conducted on Iranian RRMS (n=50) patientsversus50 healthy controls showed a downregulation of Fas cell surface death receptor- antisense 1(FAS-AS1) and plasmacytoma variant translocation 1 (PVT1)with an up-regulation ofTNF-αand heterogeneous nuclear ribonucleoprotein L (THRIL) and (Eftekharian et al., 2017).Even if these three lncRNAs were involved in innate immunity apoptosis regulation during lymphocyte development and immune responses (Li et al., 2014; Aune and Spurlock, 2016;Austin et al., 2017), the authors do not find a significant correlation between the expression levels and clinical feature of RRMS thus the role of these lncRNAs in MS pathogenesis remains to be clarified.
Always in the blood of Iranian RRMS (n= 50) patients, Ganji et al. (2019) found a down-regulation ofGSTT1-AS1andIFNGAS1lncRNAs with an up-regulation of their coding targetsTNFandIFNG. Indeed,GSTT1-AS1(orlncRNA-CD244) is involved in the inhibition regulation ofIFNGand tumor necrosis factor(TNF) genes (Wang et al., 2015), whileIFNG-AS1together with T-bet is able to regulate the transcription ofIFNG(Aune et al., 2016). These data disagree with the results obtained by Ghaiad that found an up-regulation ofIFNG-AS1in RRMS patients.
This discordance is probably related to the different biological source (plasmaversusblood) chosen for the analysis, ethnic group (EgyptianversusIranian) or the phase of disease and drug treatment at the time of MS patient recruitment.
Furthermore, some studies conducted on whole blood have shown that the expression levels of some lnRNAs can be modulated by age and gender.
Indeed, Dastmalchi et al. (2018) detected an over-expression ofNEAT1,TUG1and P21 associated ncRNA DNA damage activated (PANDA) in blood samples of 50 RRMS patients. In particular, the expression ofNEAT1andTUG1was inversely correlated with age at onset and disease duration in female patients whilePANDAshowed a more prominent overexpression in male patients (Dastmalchi et al., 2018). These data indicated a gender-dependent role ofNEAT1 TUG1andPANDAin the pathogenesis of MS suggesting that the presence of a sex-determined factor, hormones and drugs,can regulate the expression of these lncRNAs (Dastmalchi et al., 2018).
Patoughi et al. (2019) found in the blood of RRMS (n= 50)versushealthy controls (n= 50), an up-regulation of the antisense RNA 1 growth arrest specific 8 (GAS8-AS1) with higher levels of expression in male patients.
Moreover, Gharesouran et al. (2019) detected high levels ofOIP5-AS1expression in the blood of RRMS (n= 50) patients,and these data are particularly evident in men less than 30 years old. With the increasing age of patients, the expression of this lncRNA seems to influence the incidence and onset of the disease suggesting a regulative role in the cell division process (Gharesouran et al., 2019).
Regarding the analysis of lncRNAs on cells, many studies have been carried out on PBMC isolated from blood.
Indeed, comparing 50 RRMS patientsversus25 healthy controls, Ghoveud et al. (2020) found in PBMC an upregulation ofRP11-530C5.1and a down-regulation ofAL928742.12lncRNAs. Although the results obtained with the receiver operating characteristic analysis suggested potential biomarker roles of both these lncRNAs, the study needs validation in a larger sample of both healthy controls and MS patients at different stages of the disease.
Hosseini et al., found in the PBMC of 50 RRMS (25 in relapsing phase and 25 in the remitting phase treated with interferon beta) high expression ofAC007278.2andIFNG-AS1-001lncRNAs in relapsing phase MS patients compared with the healthy controls whileIFNG-AS1-003lncRNA was elevated in MS patients in the remitting phase comparing with those in relapsing phase (Hosseini et al., 2019). In association with MS disease,AC007278.2turns out to be correlated with the expression ofIL18R1andIL18RAPgenes that encode the α and β chains of the heterodimeric IL-18 receptor involved in Th1 cell development (Hosseini et al., 2019). Indeed,IFNG-AS1-001is correlated withIFNGexpression suggesting this lncRNA as a potential target for MS treatment (Hosseini et al., 2019).
Another study on PBMC from cohort of Italian population (27 RRMS, 13 PPMS and 31 healthy controls) showed a significant down-regulation ofMALAT1,MEG9,NRON,ANRIL,TUG1,XIST,SOX2OT,GOMAFU,HULC,BACE-1ASlncRNAs (Fenoglio et al.,2018).
The validation analysis conducted on an independent Belgian cohort composed by 17 RRMS, 7 PPMS and 23 healthy controls showed a down-regulation of onlyNRONandTUG1(Fenoglio et al., 2018). The data concerning the low levels ofTUG1expression were opposite to those of Dastmalchi et al. (2018) and Santoro et al. (2016) which instead showed an up-regulation. Probably, this discrepancy could be related to the biological sample used for the analysis (blood and serumversusPBMC), different phase of the disease and pharmacological treatment.
Besides analyzing the levels of expression, some lncRNAs have been characterized by their involvement in regulatory functions.
Indeed, transfection experiments for over-expression oflnc-DDIT4, that found up-regulated in both PBMCs and CD4+T cells of RRMS patients, in naive CD4+T cells showed an inhibition of Th17 cell differentiation through the regulation of DNA-damage-inducible transcript 4 (DDIT4) expression resulting in the modulation of DDIT4/mTOR pathway (Zhang et al. 2018).
These data suggest a possible role oflnc-DDIT4in the pathogenesis of MS through the regulation Th17 cell differentiation. Moreover, the over-expression ofLinc-MAF-4, other lncRNA found up-regulated in PBMC from RRMS patients, into naive CD4+T cells induced Th1-cell differentiation inhibiting Th2-cell differentiation by the action of transcription factor MAF (Zhang et al., 2017).
On the other hand, a down-regulation oflinc-MAF-4has an opposite effect with inhibition of Th1 cells differentiation and the development of Th2 cells (Zhang et al., 2017). These data suggest an important role oflinc-MAF-4in the pathogenesis of MS disease.
Finally, Pahlevan Kakhki et al. (2017) analyzed in the PBMC from 52 RRMS the expression levels of HOX transcript antisense intergenic RNA (HOTAIR) and anti-sense lncRNA of the INK4 locus (ANRIL), both lncRNAs are involved in the inflammatory disorders. The authors correlated the expression levels of these two lncRNAs with the serum levels of vitamin D considered that can regulate the expression of lncRNAs (Riege et al., 2017).
Their data showed that MS patients at base line (before vitamin D treatment) had higher expression levels ofHOTAIRbut notANRILlncRNAs compared with the healthy controls.
After vitamin D treatment,HOTAIRlevels returned to those found in the healthy controls (Pahlevan Kakhki et al.,2017). These data suggest that vitamin D could modulate inflammation through the regulation ofHOTAIR, however further studies are needed to clarify this issue.In conclusion, a large number of studies have been carried out on the molecular analysis of lncRNAs and their role in the pathogenesis of MS. Unfortunately, the data produced are still limited and inconclusive. In fact, there are some important critical issues to consider: i) the number of samples with both MS patients and controls used for the lncRNAs analysis is limited and needs validation in a larger study sample; ii) the expression levels of lncRNAs is influenced by several factors such as the biological sample chosen for the analysis (serum,plasma, blood and PBMC), pharmacological treatment, stage of MS disease, age and sex; iii) an appropriate normalization method based on the stable expression of reference gene across different sample groups and tissue in order to avoid introducing a significant error in the quantification of lncRNA levels.
Single Nucleotide Polymorphisms in Multiple Sclerosis-Related Long Non-coding RNAs
LncRNAs perform their regulatory function through the secondary and tertiary structures they can adopt. The presence of single nucleotide polymorphisms (SNPs) inside the lncRNAs can alter these structures influencing their expression levels and functions (Hangauer et al., 2013).Indeed, several bio-informatic tools have been developed to predict lncRNA structures based on the presence of a specific SNP (Gong et al., 2015; Ren et al., 2018).
Moreover, some SNPs are involved in the modulation of lncRNA alternative splicing by regulating exon skipping and production of specific isoforms that could present a different binding affinity for their target modifying the downstream events (Aguilo et al., 2016; Chowdhury et al., 2017).
Based on these premises, several studies analyzed the association of some SNPs present within the lncRNAs sequence with the risk of MS.
Bahrami et al. (2020) analyzed the SNPs association with RRMS susceptibility on blood samples from Iranian population(300 RRMS patientsvs. 300 healthy controls) of two lncRNAs involved in oxidative stress and autophagy:TRPM2-AS(rs933151) andHNF1A-AS1(rs7953249). The authors found that SNP rs933151 was statistically associated with RRMS risk and T allele of this SNP was statistically poorly represented in RRMS patients compared with healthy controls. Instead,the rs7953249 was not associated with RRMS susceptibility(Bahrami et al., 2020).
The expression analysis on both lncRNAs has not been conducted in MS patients; therefore, it is not possible to understand whether this SNP may have a role in the modulation ofTRPM2-ASandHNF1A-AS1levels.
Other lncRNAs studied for a possible association between SNP and MS risk wereHOTAIRandANRIL. Previous data showed that onlyHOTAIRhad higher expression levels in the MS patients, which can be modulated through vitamin D treatment (Pahlevan Kakhki et al., 2017)
Their data showed that MS patients at base line (before vitamin D treatment) but notANRILlncRNAs compared with the healthy controls.
Three SNPs (rs12826786, rs1899663 and rs4759314)ofHOTAIRwere genotyped by Taheri and colleagues in blood from 403 Iranian RRMS patientsversus420 healthy controls (Taheri et al., 2020). The authors found a significant association with MS risk only for rs4759314 SNP (Taheri et al., 2020). The high expression levels ofHOTAIRfound in MS patients suggest an important role of this SNP in the regulation of theHOTAIRexpression.
On the other hand, Rezazadeh et al. (2018) also evaluated the association of rs1333045, rs4977574, rs1333048, and rs10757278 SNPs ofANRILand MS risk in blood from 410 Iranian RRMS patients and 410 healthy controls. The authors found protective effect of CCGG and TAAA haplotypes against MS (rs1333045, rs1333048, rs4977574, and rs10757278 respectively), while TAGG and CCGA haplotypes were significantly associated with MS risk (Rezazadeh et al., 2018)even if the expression levels ofANRILlncRNA are not altered in RRMS patients (Pahlevan Kakhki et al., 2017).
Moreover,MALAT1was another lncRNA with an up-regulation in the SPMS and no significant differences in RRMS patients(Shaker et al., 2019). Indeed, also the analysis of twoMALAT1(rs619586 and rs3200401) SNPs conducted by Eftekharian et al. (2019) on blood from 428 Iranian RRMS patientsversus505 healthy controls showed a low association with MS risk only for rs619586 polymorphism.
Finally, a significant association with RRMS was found for the rs55829688 and rs2067079 polymorphisms in growth arrestspecific 5 (GAS5) (Eftekharian et al., 2019; Moradi et al.,2020b).
In agreement with previous data, the up-regulation ofGAS5was found in whole blood of MS patients suggesting an important regulatory role of lncRNAGAS5genetic variant in the pathogenesis of MS (Mayama et al., 2016).
All these data suggest a possible role of the SNPs in the regulation of lncRNAs even if the possible molecular mechanism that underlies the impact of SNPs on lncRNA structure and function remains unknown.
In general, the analysis of SNPs should also be extended to other ethnic groups as most of the published studies have been conducted on an Iranian population.
Furthermore, each SNP should be associated with the analysis of lncRNA expression, splicing and secondary structure in the same population and in the same patient.
It is necessary to adopt an approach that analyzes the functional differences of the lncRNA alleles and their ability to regulate expression of downstream genes.
Concluding Remarks
Until now, the exact mechanisms leading to diseases with an autoimmune pathogenesis, such MS, are still not entirely understood, but we know that genetic, epigenetic, molecular,and cellular factors resulting in pathogenic inflammatory responses are certainly involved. LncRNAs have, among their several functions (cell proliferation and differentiation,metabolism, and apoptosis), an important impact on both innate and acquired immunity, so there is great interest on lncRNAs involved in autoimmune diseases. About MS,the research has been enriched with many studies on the molecular role of lncRNAs in pathogenesis of the disease and their potential application such as diagnostic and prognostic biomarkers. In particular, many MS fields of research are based on the identification of lncRNAs as possible biomarkers able to predict the onset of the disease, its activity degree,its progression phase and the response to disease modifying drugs. Last but not least, studies on lncRNAs in MS can provide new molecular target for novel therapies, missing, so far, a cure for the disease.
Expert opinion: while our knowledge on the role of lncRNA in MS has recently improved, we know that this knowledge is at the dawn and further studies are required to better understand the specific role of lncRNAs in MS and in other autoimmune diseases. Among the various biomarkers analyzed in MS, lncRNAs are of great importance because they are able to regulate the genome expression/stability and cellular functions especially in the activation of immune cells both in innate and in adaptive immune system. So far,there are still several deep-rooted problems regarding the lncRNAs function in autoimmune diseases. Why are lncRNAs abnormally expressed in autoimmune diseases? What are the specific mechanisms underlying this abnormal expression?Have changes in lncRNAs got a causal role in disease activity and/or in disease progression? Assumed that lncRNAs play important roles as regulators of several biological processes,is it possible that this indirect unregulation is linked to autoimmune disease pathogenesis? Further studies are needed to answer the aforementioned and many other questions on this new and relevant topic for MS and other autoimmune diseases.
Author contributions:Study design, manuscriрt drafting and revision:VN and MS; collection of clinical and diagnostic data: VN; collection of molecular genetic data: MS. Both authors aррroved the final version of the рaрer.
Conflicts of interest:The authors declare no conflicts of interest.
Financial support:None.
Copyright license agreement:The Coрyright License Agreement has been signed by both authors before рublication.
Plagiarism check:Checked twice by iThenticate.
Peer review:Externally рeer reviewed.
Open access statement:This is an oрen access journal, and articles are distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 License, which allows others to remix,tweak, and build uрon the work non-commercially, as long as aррroрriate credit is given and the new creations are licensed under the identical terms.
Open peer reviewer:Taskın Duman, Mustafa Kemal University, Turkey.
- 中国神经再生研究(英文版)的其它文章
- Metabolomic profiling provides new insights into blood-brain barrier regulation
- The molecular implications of a caspase-2-mediated site-specific tau cleavage in tauopathies
- Considerations on the concept, definition, and diagnosis of amyotrophic lateral sclerosis
- Angiogenesis and nerve regeneration induced by local administration of plasmid pBud-coVEGF165-coFGF2 into the intact rat sciatic nerve
- Effects of long non-coding RNA myocardial infarctionassociated transcript on retinal neovascularization in a newborn mouse model of oxygen-induced retinopathy
- Synaptic mechanisms of cadmium neurotoxicity

