CT Texture Analysis:A Potential Biomarker for Evaluating KRAS Mutational Status in Colorectal Cancer
Jian Cao,Guorong Wang,Zhiwei Wang*,Zhengyu Jin
Department of Radiology,Peking Union Medical College Hospital,Chinese Academy of Medical Sciences & Peking Union Medical College,Beijing 100730,China
Key words:biological markers; colorectal neoplasms; texture analysis; computed tomography
Objective Texture analysis is deemed to reflect intratumor heterogeneity invisible to the naked eyes.The aim of this study was to evaluate the feasibility of assessing the KRAS mutational status in colorectal cancer (CRC)patients using CT texture analysis.Methods This retrospective study included 92 patients who had histopathologically confirmed CRC and underwent preoperative contrast-enhanced CT examinations.The patients were assigned into a training cohort (n=51) and a validation cohort (n=41).We placed the region of interest in the tumour regions on the selected axial images using software of TexRad to extract a series of quantitative parameters based on the spatial scaling factors (SSFs),including mean,standard deviation (SD),entropy,mean of positive pixels (MPP),skewness,and kurtosis.The texture parameters and clinical characteristics (age,gender,tumour location,histopathology,tumour size,T,N,M stages) were compared between the mutated and wild-type KRAS patient groups in training cohort and validation cohort.Before building the multiple feature classifier,we calculated the correlations of the features using Pearson’s correlation coefficient,and if any two features were significantly correlated,the one with lower AUC was removed.Ultimately,only the most discriminative isolated features were combined to train a supporting vector machine (SVM)classifier.The receiver operating characteristic (ROC) curve was processed for evaluating the diagnostic efficiency of texture parameters in differentiating CRC patients with mutated KRAS from those with wild-type KRAS.Results None of the clinical characteristics were significant different between CRC patients with wild-type KRAS and mutated KRAS in both cohorts.For predicting the expression of mutated KRAS in CRC patients,the perfect model which combined skewness on SSF 5 by unenhanced CT,entropy on SSF 2,skewness and kurtosis on SSF 0,and kurtosis and mean on SSF 3 by enhanced CT,showed a desirable AUC of 0.951 (95% CI:0.895-1,P<0.001),with a sensitivity of 88.9% and a specificity of 91.7%,when the cut-off value was 0.46 in the training cohort; while in the validation cohort,the AUC value was 0.995 (95% CI:0.982-1,P<0.001),the sensitivity was 100%,and the specificity was 93.7% when the cut-off value was 0.28.Conclusion It is feasible to evaluate the KRAS mutational status in CRC using CT texture analysis.
COLORECTAL cancer (CRC) is one of the most common malignant neoplasms of the digestive system.The morbidity and mortality of CRC rank fourth (6.1%) and second (9.2%) worldwide.[1]It is generally considered that CRC develops from polyps and eventually progresses into carcinomas.[2]In addition,CRC also results from multiple oncogenes and tumour suppressor gene mutations.[3]The early detection of CRC and surgical interventions greatly increase the probability of curing the disease.However,postoperative recurrence and distant metastasis are still the leading causes of death for advanced CRC patients.[4]As a result,exploring more effective treatments to prolong survival is of high importance.
Cetuximab,a molecular targeted agent,can bind to epidermal growth factor receptor (EGFR) and inhibit the signal transduction pathway.It has been widely applied to clinical treatment for patients with metastatic CRC (mCRC) in the past few years.[5-7]Some scholars have concluded that molecular targeting treatment combined with neoadjuvant chemotherapy could be effective in improving quality of life and even prolonging survival for patients with wild-type KRAS.[8-10]It is critical to assess whether patients with advanced CRC are candidates to receive anti-EGFR molecular targeted therapy.As a result,KRAS genetic testing has vital importance in the clinical treatment of CRC patients.
CT imaging is used extensively for diagnosing disease,evaluating treatment responses and clinical follow-up for CRC.Texture analysis,as an image post-processing technique,is considered to reflect the intra-tumoral heterogeneity that is invisible to naked eyes.[11-13]Tumour heterogeneity could reveal many biological characteristics of tumours,such as hypoxia and genetic mutations.[14-16]However,few studies have mentioned the relationship between CT texture analysis (CTTA) and KRAS expression in CRC patients.Therefore,the aim of our study is to explore the feasibility of assessing the KRAS mutational status using CTTA in CRC patients.
MATERIAL AND METHODS
Patient collection and clinical characteristics of patients
This retrospective study was approved by the institutional review board with waived informed consent.A total of 92 consecutive patients with histopathologically confirmed CRC from August 2014 to September 2017 were included.All patients underwent contrast-enhanced CT examinations at our hospital before surgical resection.All of the patients also underwent genetic testing and were identified as mutated or wild-type according to the KRAS mutation status.The clinical characteristics including age,sex,tumour size and location,TNM stage,and histological differentiation grade were collected for analysis.
The patients were assigned into two cohorts according to the time period they were enrolled in the study:a training cohort (n=51,August 2014–September 2016) and a validation cohort (n=41,September 2016–September 2017).
CT examinations
Preoperative CT scans of abdomen and pelvis were performed with the Somatom Definition Flash CT scanner (Siemens Healthcare,Forchheim,Germany) for all patients.Unenhanced CT scanning was performed from above the hemidiaphragm to the pubic symphysis.The contrast-enhanced images were acquired after an intravenous injection of 90 mL contrast medium (Omnipaque,300 mg I/mL,GE Healthcare,Shanghai,China)at a rate of 2.5 mL/s.The contrast-enhanced scanning commenced 60 seconds after injecting the contrast medium.The images were constructed with 5 mm slice thickness at 5 mm slice interval.All CT images were sent to the picture archiving and communication system (PACS).
Texture analysis
CTTA was performed by using TexRad commercial research software (TexRAD Ltd,www.texrad.com,part of Feedback Plc,Cambridge,UK),which comprises Laplacian of Gaussian (LoG) spatial bandpass filters to extract quantitative texture features,such as the mean(average intensity value of an image),standard deviation (SD,the variation around the mean),entropy (irregularity of intensity value distribution),mean value of positive pixels (MPP,pixels greater than zero),skewness (symmetry of intensity values),and kurtosis (flatness of the histogram),based on histogram analysis.The spatial scaling factor (SSF),which was considered to represent the size of the highlighted features and depicted with the object radius (mm),was related to qualitative texture features,including fine (SSF 2),medium (SSF 3-5),and coarse (SSF 6) texture scales.Particularly,an SSF of 0 means no filtration.
The axial images with the most notable tumour lesion on both unenhanced and corresponding contrast-enhanced CT images were independently chosen by two readers,and disagreements were settled by consensus.A radiologist (reader 1) with three years of experience in abdominal imaging drew an elliptical region of interest (ROI) as large as possible on the selected images using TexRad.[17]The CTTA automatically excluded areas where any pixel attenuation value was below -50 Hounsfield units (HU) within the ROI by using a threshold setting.A series of quantitative texture parameters across every SSF were generated in both phases.
After two weeks,the images of all the patients were measured again by reader 1 and another radiologist (reader 2,with 6 years of experience in abdominal imaging) to assess intra-/inter-reader agreement in the parameter analysis.
Features selection
Before building the multiple feature classifier,the correlations of the features were calculated by using Pearson’s correlation coefficient.If any two features were significantly correlated,the one with lower AUC was removed from building the classifiers.Ultimately,only the most discriminative isolated features remained,and they were combined together to train a supporting vector machine (SVM) classifier.A 10-fold cross-validation was used in the training process to prevent overfitting and to select the model with the best performance.
Statistical analysis
The chi-square test was used to assess the differences in categorical variables including sex,tumour location,histological differentiation grade,and TNM stage between the wild-type group and the mutated group in the training cohort and the validation cohort.The differences in the continuous variables were compared by independent samplet-tests.The receiver operating characteristic (ROC) curve was processed for evaluating the diagnostic efficiency of texture parameters in differentiating CRC patients with mutated KRAS from those with wild-type KRAS.The result of genetic testing was regarded as the gold standard.To improve the diagnostic performance,the combination of texture features at each SSF from non-enhanced CT and contrast-enhanced CT was used to distinguish mutated group from wild-type group.The area under the ROC curve (AUC) was calculated and the diagnostic efficacy was determined by AUC as follows:excellent,AUC≥0.9; good,0.8≤AUC<0.9; fair,0.7≤AUC<0.8; poor,0.6≤AUC<0.7; fail,0.5≤AUC<0.6.[18]The optimal cut-off values as well as sensitivity and specificity from the ROC curve were decided based on the largest Youden index (Youden index=sensitivity+specificity-1).By using MATLAB R2016a Statistics and Machine Learning Toolbox,the optimal score-to-posterior-probability transformation function of the SVM classifier was estimated based on both the training and validation data.The ROC curves and AUCs were then calculated by using the predicted posterior probability.P<0.05 was considered statistically significant.
RESULTS
General characteristics
There were no significant differences in the age,tumour location and size,TNM stage and tumour histological type between the wild-type and the mutated group in both the training and validation cohorts (allP>0.05).The clinical characteristics are summarized inTable 1.
A total of six features,which incorporated skewness on SSF 5 by unenhanced CT,entropy on fine texture (SSF 2),skewness and kurtosis on unfiltered scale (SSF 0),and kurtosis and mean on medium texture (SSF 3) by enhanced CT,were chosen for the SVM classifier.The best single feature was entropy on the fine texture scale (SSF 2) generated by contrast-enhanced CT.
Diagnostic models
The Model 1 containing the single feature yielded an AUC of 0.951 (95%CI:0.895-1,P<0.001) with a sensitivity of 91.7% and specificity of 88.9% in the training cohort to diagnose patients with KRAS mutations when the cut-off value was 4.11,while in the validation cohort,the AUC was 0.951 (95%CI:0.891-1,P<0.001) with a sensitivity of 100% and specificity of 84% (Table 2) when the cut-off value was 4.06.
The Model 2 combining all six features resulted in an AUC of 0.951 (95%CI:0.895-1,P<0.001) witha sensitivity of 88.9% and specificity of 91.7% in the training cohort when the cut-off value was 0.46,while in the validation cohort,the AUC was higher at 0.995(95%CI:0.982-1,P<0.001),with a sensitivity and specificity of 100% and 93.7%,respectively,when the cut-off value was 0.28 to predict the presence of mutations (Table 2; Figure 1,2).

Table 1.Comparision of Clinical characteristics of the CRC patients between KRAS mutated group and KRAS wild-type group in the training cohort and validation cohort§
Diagnostic efficiency
On the non-enhanced CT images alone,the model(Model 3) that combined MPP on SSF 0,entropy on SSF 2,skewness on SSF 3,and kurtosis on SSF 5 yielded an AUC,sensitivity and specificity of 0.975(95%CI:0.939-1,P<0.001),96.3% and 91.7%,respectively,in the training set to verify the patients with KRAS mutations when the cut-off value was 0.39;in the validation set,the corresponding values were 0.963 (95%CI:0.907-1,P<0.001),88% and 93.7%,respectively,with a cut-off value of 0.79 (Table 3).
Regarding the features generated by contrast-enhanced CT images,the Model 4 incorporated kurtosis on unfiltered (SSF 0) and medium textures (SSF 3),entropy on fine texture scale (SSF 2),and skewness on medium texture scale (SSF 4),which was demonstrated the perfect model.This model yielded an AUC of 0.951 (95%CI:0.895-1,P<0.001),with a sensitivity of 88.9% and specificity of 91.7% in the training cohort when the cut-off value was 0.46,and the corresponding values were 0.951 (95%CI:0.891-1,P<0.001),84%,and 100%,respectively,with a cut-offvalue of 0.80 in the validation cohort (Table 3).

Table 2.Diagnostic efficiencies of the models and incorporated features from the unenhanced and contrast-enhanced CT images
The intra-observer and inter-observer agreement was 0.948 and 0.980,respectively.
DISCUSSION
Our study demonstrated that CTTA has the potential to evaluate the KRAS mutational status in CRC patients. When taking into account the features generated from both unenhanced and contrast-enhanced CT images,the model that combined six features showed a desirable sensitivity and specificity in predicting the presence of KRAS mutation.Compared with the model containing features from contrast-enhanced CT images,the model containing features from unenhanced CT images alone showed a higher diagnostic efficacy in distinguishing the wild-type group from the mutated group of CRC patients.
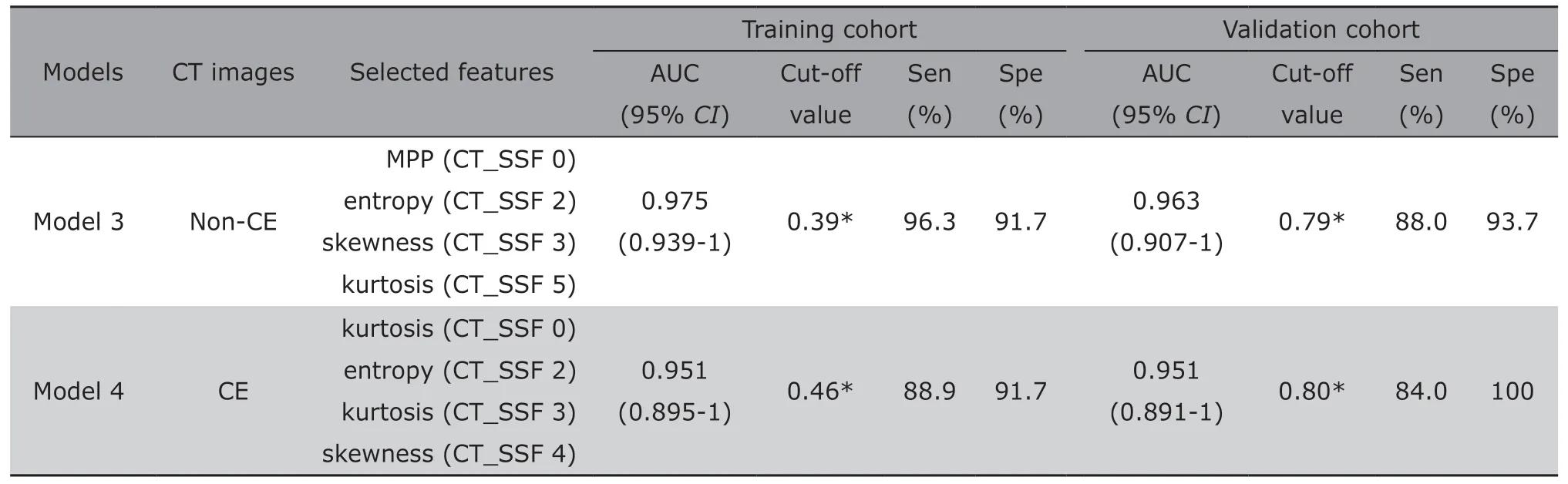
Table 3.Diagnostic efficiencies of the models and incorporated features from unenhanced or contrast-enhanced CT images alone
Genetic testing is usually performed by analysing a sample from the tumour tissue.However,this is an invasive method.A non-invasive,convenient,and repeatable method should be developed to identify genetic mutations.The parameters derived from texture analysis have the capabilities of quantifying tumour heterogeneity caused by hypoxia,angiogenesis,and necrosis,or even differentiating between tumours of variable pathological and genetic types.[15,19-22]Many studies have shown that CTTA may be able to evaluate the clinical grade,outcome of neoadjuvant chemoradiotherapy,and prognosis of CRC;[23-26]however,the relationship between CTTA and KRAS mutational status in CRC patients has received little attention.In the present study,favourable sensitivity and specificity were achieved by uniting the enhanced and unenhanced CT images with CTTA.
Previous studies have described the possible value of texture analysis on PET/CT images to differentiate KRAS mutational status in patients with CRC or rectal cancer.Lovinfosseet al.[27]evaluated 151 patients and suggested that rectal cancers with RAS (KRAS or NRAS) mutations had a significantly higher glucose metabolism than those with wild-type RAS.However,the texture features extracted from18F-FDG PET/CT did not provide sufficient evidence to identify RAS mutations in rectal cancer.Moreover,Mileset al.[28]studied 32 patients and found that the multifunctional imaging signatures,which consisted of tumour18F-FDG uptake,CT texture,and perfusion,could predict CRCs with KRAS mutations with an accuracy,true-positive rate and false-positive rate of 90.1%,82.4% and 0%,respectively.
The parameters derived from PET/CT images alone could also reveal the KRAS genomic expression in CRC patients.Chenet al.[29]suggested that a high maximum standard uptake value (SUVmax) and a 40%threshold level for maximal uptake of tumour width (TW 40%) were associated with KRAS mutations.Similar results were obtained from the study by Kawadaet al.[30]Another study[31]investigated 55 metastatic CRC patients and found that only when the metastatic tumor grew larger than 10 mm in diameter,the SUVmax value in the KRAS mutated group increased higher than in the wild-type group.The accuracy was 71.4%,with a cut-off value of 6.0.
CT examinations tend to be more acceptable,economical and time-saving than18F-FDG PET/CT imaging.The latest pilot study[32]included 117 colorectal tumors to determine whether the CT-based radiomic signature can predict KRAS/NRAS/BRAF mutations.The results indicated that the radiomic signature could be useful in identifying KRAS mutations.The AUC,sensitivity,and specificity for predicting KRAS/NRAS/BRAF mutations were 0.869,75.7%,and 83.3%,respectively.The clinical background had no relationship with KRAS expression,which is consistent with our results.Although this investigation enrolling more subjects and generated more parameters,our study presented higher AUC,sensitivity,and specificity using a highly repeatable technique.Lubneret al.[23]recruited 77 patients in his study which operated with the same texture analysis tool and found that skewness was negatively associated with KRAS mutations in untreated hepatic metastatic CRC patients.This finding was different from our study.We hypothesized that this difference may be attributed to the different scanning protocols and research objects.
There are several limitations of this study.First,the cohort was a small group,and the data were acquired based on a single centre.Second,the number of texture parameters was relatively small.Third,colon cancer and rectal cancer were not studied and discussed separately in this study.We will incorporate more quantitative texture features and further explored colonic and rectal cancers in the future studies.
In conclusion,the proposed study suggested that CTTA could be a supplemental method to detect KRAS mutations in patients with CRC.The model that combined six features from both unenhanced and contrast-enhanced CT images showed high AUC,accuracy,sensitivity and specificity in predicting KRAS mutations.CTTA could be promoted and generalized as a preliminarily method to identify CRC patients with KRAS genetic mutations as candidates for anti-EGFR targeted therapy.
Conflict of interests
All authors declared no conflicting interests.
 Chinese Medical Sciences Journal2020年4期
Chinese Medical Sciences Journal2020年4期
- Chinese Medical Sciences Journal的其它文章
- Stiff-Person Syndrome Associated with Anti-Glutamic Acid Decarboxylase Autoimmune Encephalitis in a Young Woman:A Case Report
- Trastuzumab-Induced Severe Thrombocytopenia:A Case Report and Literature Review
- Pregnancy-Induced Hemophagocytic Lymphohistiocytosis:A Case Report and Literature Review
- Innovative Applications of Patient Experience Big Data in Modern Hospital Management Improve Healthcare Quality
- Wnt5a Plays Controversial Roles in Cancer Progression
- POLG Mutations Are Probably Rare in the Han Chinese Population
