Potential of transposon-mediated cellular reprogramming towards cell-based therapies
Dharmendra Kumar, Taruna Anand, Thirumala R Talluri, ilfried A Kues
Dharmendra Kumar, Animal Physiology and Reproduction Division, ICAR-Central Institute for Research on Buffaloes, Hisar 125001, India
Taruna Anand, NCVTC, ICAR-National Research Centre on Equines, Hisar 125001, India
Thirumala R Talluri, Equine Production Campus, ICAR-National Research Centre on Equines, Bikaner 334001, India
Wilfried A Kues, Friedrich-Loeffler-Institut, Institute of Farm Animal Genetics, Department of Biotechnology, Mariensee 31535, Germany
Abstract
Key words: Transposons; Induced pluripotent stem cells; Clinical applications; Cellular reprogramming; Cell-based therapy; Genetic correction
INTRODUCTION
Transposon systems currently provide a promising toolbox for cell therapy, disease modeling, and drug discovery[1-4].Importantly, the non-viral transposon systems can be an important alternative to viral vectors, which are commonly used for cellular reprogramming for transfection of somatic cells with exogenousOct4,Sox2,Klf4, andc-Mycgenes to induce cellular pluripotency and establish induced pluripotent stem (iPS) cells[5-8].However, the limited cargo size of retro and lenti viral vectors of about 7 kb pairs hampers transfer of larger therapeutic genes[9].In addition, the construction of viral vectors is cumbersome, expensive and requires living cells for their scale up, which further complicates the quality control and downstream processing[10].
The iPS cell technology promises to provide an unlimited source of cells for innovative therapies, and to treat so far incurable diseases[11-13].A hypothetical schedule would require a small tissue sample from the patient, to reprogram the somatic cells to iPS cells with unlimited proliferative capacity, to perform gene correction in the iPS cells, then to direct differentiation into the desired precursor cells, which are finally transplanted into the patient (Figure 1).
In this respect, Sleeping Beauty (SB) and piggyBac (PB) transposon systems appear as attractive tools for somatic cell reprogramming due to their efficient gene delivery and their ability to be excised from the cells after reprogramming, which helps overcome the limitations of viral-based reprogramming technologies.Transposon systems have a number of additional advantages, such as (1) Cargo capacity of up to 100 kb[14,15]; (2) No bias to integrate in expressed genes or promoter regions; (3) Possibility of seamless removal of the transposon[16,17]; (4) Cost-effective production of the basic plasmids; (5) Reduced innate immunogenicity; and (6) No requirement for a specialized biosafety facility.
The translation of this iPS cell-based therapy into clinical testing needs authorization approval to initiate safety and efficacy studies, and to exclude risks of insertional oncogenesis or immunogenicity[18,19].SB and PB transposon systems have been successfully used to obtain reprogrammed iPS cells from human somatic cells[16,20], but also somatic cells from the murine model[21-24], and cells from large model species, such as pig[25], horse[26], bat[27], monkey[28], rat[29], cattle[30,31]and buffalo[32].Here, we review the potential of transposon-mediated cellular reprogramming and its clinical applications in cell-based therapy and the associated risks.
SHORT SYNOPSIS OF THE MOST COMMONLY APPLIED TRANSPOSON SYSTEMS
DNA transposons, also known as Class II elements or mobile genetic elements, were first described as “jumping genes” by McClintock[33]and were found to be responsible for color mosaicism of maize cob kernels.DNA transposons have been divided into two major groups: (1) Cut-and-paste; and (2) Rolling-circle transposons[34].In vertebrates, commonly cut-and-paste group of transposons are found, which include the Tc1/mariner, hATs, PB and SB families, all of which are characterized by inverted terminal repeats of 10 to 1000 bp flanking their transposase gene[35].Transposons are discrete DNA segments which can move from one site to another within a genome, and sometimes between genomes catalyzed by the transposase[36,37].Transposons are species-specific, found in the genomes of all prokaryotes and eukaryotes, whereas in humans approximately 46% of the genome is derived from retro- (RNA) and DNA transposons[38,39].

Figure 1 Schematic representation of induced pluripotent stem cell derivation, differentiation and genetic modification.
Transposons are important sources of genome structures that are actively used to regulate the multicellular embryonic development.These structures include binding sites with transcription factors, enhancers and silencers, promoters, insulators, alternative splicing sites, and non-coding RNA.Moreover, transposons are involved in the emergence and evolution of new protein-coding genes through exonization, domestication, and the formation of retrogenes.The activation of transposons is needed to regulate the differentiation and reproduction of cells in the body; however, in terminally differentiated cells, upon reaching predetermined sizes of organs, molecular systems are activated that block a further cascade of transposon activation[40,41].Due to the wide distribution and diversity of transposons, they contribute significantly to genomic variation and as such, they are powerful drivers of genome evolution[36,42-45].
For this purpose, SB and PB transposon systems are identified as efficient vectors for cellular reprogramming.The SB originated from salmonid fish species, where it existed as an inactive element[46]; from this a synthetic transposon system was constructed using a reverse engineering approach to eliminate the accumulated mutations[46].PB was derived from an active element discovered in the mothTrichoplusia ni[47].These transposons have no orthologous elements in mammalian species, which prevents the re-mobilization of transposons by potential endogenous transposases.This has been experimentally verified in transgenic mice and pigs[48,49].Presently, the hyperactive versions of SB (SB100X) or PB (hypPB) seem to be the most active transposon systems.They possess comparable activity levels in mammalian cells, and are independent of cellular co-factors[50,51].Both of these transposons have been employed for stable expression of reprogramming factors and are suitable for the derivation of iPS cells as proven in various studies[16,22,23,25,26,30-32,52].Other transposons namely: Frog Prince, Mos1, Tol2 and Passport are also active in mammalian cells, but they are still under-investigated in iPS cell generation[53].
MECHANISM OF TRANSPOSON-MEDIATED CELLULAR REPROGRAMMING
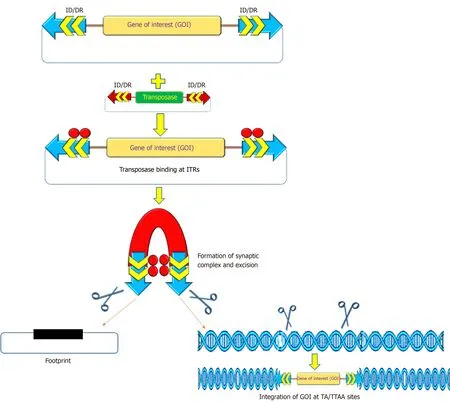
Figure 2 Mechanism of action of transposon-transposase mediated transposition.
The recombinant PB and SB systems mobilize or transfer gene(s) of interest through a “cut and paste” mechanism (Figure 2)[2,54,55].For most applications, recombinant transposon systems encompass a donor plasmid that carries one or more genes flanked by the inverted terminal repeats (ITRs) sequences essential for transposition[2,56,57].The transposase gene can be positioned on a separate plasmid (trans) or in the same plasmid (cis).Once the transposase protein is expressed, it binds to the ITR sequences, which catalyzes the removal of the gene of interest (cut) and integrates (paste) the transposon sequence into the genome of a host cell[57].The SB transposase catalyzes integrations at consensus TA-dinucleotides[46], whereas the PB requires TTAA-tetranucleotide sequences[58-60].The efficiency of transposition of these transposon systems has been further increased due to generation of highly active and efficient transposases, namely hyp(er) PB (hypPB) and hyperactive SB 100X (hySB100X)[50,51,61,62].The hySB100X showed a 30% higher transposition rate compared with SB100X.hySB100X was obtained by mutation in short hydrophilic residues in the catalytic domain of the SB100X transposase molecule, which required direct DNA contact to increase the DNA binding affinity of the transposon[62].Furthermore, the transposition rate of these transposons is affected by topological conformations, chromatin condensation and CpG-methylation patterns of the target DNA[63,64].Genomic insertion for SB100X prefers target regions with higher AT content, in a palindromic core unit[65,66]; whereas PB transposase integration requires a TTAA recognition sequence and exhibits a bias toward insertions in genes[67].
For cellular reprogramming, the transfection of the transcription factors into somatic cells using the transposon system is relatively straightforward.The transposonsmediated cellular reprogramming leads to an overall efficiency of approximately 0.02%[20,22,23,30], which nears the initially obtained reprogramming efficiencies by viral vectors.The obtained reprogramming efficiency from transposons is higher than other reported non-integrative delivery systems including either replicating episomal vectors or minicircles[68,69], although lower than Sendai viral vectors or synthetic mRNA[70,71].Transposons-mediated transposition is a self-regulated activityviaoverproduction inhibition, a mechanism by which transposition activity is downregulated when the transposase is over concentrated in cells[72].Ideally, the transposase is expressed only for a short period, which prevents continuous transposon remobilization.However, it is also important to minimize the number of vector copies per cell as it poses an increased risk of insertional oncogenesis[73].
THE EXPANDING TRANSPOSON TOOLBOX
Transposon systems are widely used for gene delivery applications[58,74-76].However, like the lenti viruses, transposon vectors are mutagenic, because of their random integration.Recently, clustered regularly interspaced short palindromic repeats (CRISPR) and Cas9 nucleases have emerged as excellent tools for site-specific mutation of genomes[77].This system is an attractive candidate for targeting through extensive base pairing with the target[78].In contrast, most DNA binding proteins remain bound to their target sites only for a matter of seconds or minutes.However, double-stranded breaks induced by CRISPR-Cas9 nucleases showed undesirable outcomes in terms of large deletions extending over many kilobases at high frequency and complex genomic rearrangements[79].To overcome the challenges of nuclease-based gene delivery, various research groups have attempted to use site-specific DNA binding proteins such as SB, PB, Mos1, and ISY100-fused with zinc finger protein, transcription activator like effector (TALE) and/or Gal4 to target specific loci[80-82].Owenset al[83]fused a TALE DNA-binding domain (DBD) with PB to direct the transposase to stimulate insertional activity of PB at the intended target sequence.This approach allowed the isolation of clones harboring single-copy insertions at the CCR5 locus.Subsequently, attempts were made using catalytically dead Cas9 (dCas9) for targeting PB insertions to the human endogenous hypoxanthine phosphoribosyl transferase (HPRT) locus[82].Surprisingly, the dCas9-PB chimera protected it from insertions instead of targeting the HPRT locus.Although, PB is considered to be the most efficient system for gene deliveryin vivo[84,85], it impedes the development of advanced applications such as direct delivery of transposons[86].To resolve this difficulty, Chen and Wang described a Cas-Transposon (CasTn) system for genomic insertions which uses a Himar1 transposase fused with a dCas9 nuclease to mediate programmable, site-directed transposition[87].They demonstrated that the Himar–dCas9 fusion protein improved the frequency of transposon insertion at a single targeted TA dinucleotide by > 300-fold compared to the un-fused transposase.This work highlights CasTn as a new modality for host-independent, programmable and site-directed DNA insertions[87].
More recently, Hewet al[88]tested a group of RNA-guided transposase vectors comprising mutations in the native PB DBD for their ability to target a single sequence in theCCR5gene.This RNA-guided transposition in human cells might be a framework for improved targeting vectors with potential applications in gene therapy and genome editing research[88].Similarly, Steckeret al[89]found that the CRISPRassociated transposase derived fromScytonema hofmanni(ShCAST), catalyzes the sitespecific RNA-directed unidirectional integration and is located a fixed distance to one side of the targeted DNA site.These sequence-specific integrations offer significant advantages over traditional virus-based integrating vectors by avoiding insertion into unwanted regions[90-93].Another approach applied to generate “transient transgenesis” by mutation at position 248 in the SB transposase to gain further insight into the transposition mechanism and for the generation of reprogramming factor-free iPS cells[17].The amino acid present at position 248 of the SB transposase is involved in an interaction with target DNA, and because of the absence of integration activity, transposon removal by these transposase mutants results in extra-chromosomal circles, thereby terminating the transposition reaction[17,94].This indicates that by the switching of a single amino acid, the SB transposase has into efficient unidirectional removal ability with utility in cellular reprogramming.In addition, soluble variants of the SB protein have been developed by genetic engineering, which allows for more control over the exposure time[95].These underlying genome engineering procedures will reduce costs and improve the safety of genome modifications.
TRANSPOSON-MEDIATED CELLULAR REPROGRAMMING
Commonly, somatic cells were reprogrammed to pluripotency by the exogenous introduction of transcription factors (Oct3/4, Sox2, Klf4 and c-Myc).The resulting iPS cells demonstrate the features of embryonic stem (ES) cells, including the ability to form chimeras and contribute to the germ line[5].Thereafter, iPS cells were generated either by the protein transduction approach[96], or in combination with small chemical molecules[97]without genetic modification.These reprogramming approaches suffer from low efficiency and require complicated and prolonged cell culture conditions[96,97].Furthermore, these approaches need either extraction of crude cell lysates of cells expressing defined reprogramming factors or preparation of a large amount of recombinant reprogramming transcription factors from bacteria, which may be contaminated with unknown detrimental genetic materials.Thus, the use of a suitable gene-delivery reprogramming approach is a critical step in the generation of iPS cells for basic and clinical research.
More recently, DNA transposons appeared as alternative tools for cellular reprogramming in a wide range of cell types, including fibroblasts using cocktails of transcription factors.This technique is straightforward, less time consuming and easy to handle as compared to viral vectors (Figure 3).In general, PB and SB systems have been used for iPS cells generation in a broad range of domesticated and farm animal species[16,20,22,23,25,30,32,98-101], in addition to human cells[102-105].The generation of iPS cells from domesticated and companion animal species such as cattle, pig, horse and buffalo is critically important for the establishment of disease models and economically valuable for the production of medically useful substances,e.g., enzymes and growth hormones, which are either absent or inadequate in patients suffering from specific genetic diseases.More importantly, either iPS cells or differentiated cells from iPS cells could be directly used for cellular therapies, drug screening, and disease modeling thus significantly decreasing the extent to which animals are used for research purposes[4,106-110].
In this direction, cellular reprogramming through transposon systems represents one of the unique features of the excision of gene expression cassettes from the iPS cell genome through re-expression of integration-deficient transposase variants.Alternatively, excision can be achieved by either clustered regularly interspaced short palindromic repeats/CRISPR-associated protein-9 (CRISPR/Cas9) or Cre/loxP recombination technology[22,94].Using these technologies enable the production of “transgene-free” iPS cells, which could be beneficial in minimizing the risk of reactivation of reprogramming factors leading to oncogenic potential[94].Similarly, Woltjenet al[111]showed that PB-mediated transgene excision does not leave a genetic trace in the host genome, thus providing the feasibility of seamless modification for “genetically unmodified iPS cells” production.
DIFFERENTIATION POTENTIAL OF TRANSPOSON-MEDIATED IPS CELLS
Currently iPS cells are considered a valuable resource for studying medicine and regenerative biology due to their tremendous differentiation capacity into almost all cell types of the body.In principle, the differentiated cells derived from iPS cells should behave in the same way as theirin vivocounterparts in terms of both molecular and functional aspects, but it remains a challenge to direct cell fate decisions underin vitroconditions towards specific cell types[112].In general, differentiation comprises the conversion of an iPS cell to a more specialized cell type, involving a transition from proliferation to specialization.This involves successive alterations in cell morphology, membrane potential, metabolic activity and responsiveness to specific signals.Differentiation leads to acquiring specific functions of differentiated cells depending upon the tissue in which they will finally reside[113].
The transposon-mediated iPS cells can be differentiatedin vitroin the absence of appropriate growth factor (LIF/bFGF) or feeder cells.Under the appropriate conditions, such as suspension culture, embryoid bodies (EBs) can be formed from iPS cells of almost all species, such as human[20], mouse[21,23], bat[27], monkey[28], prairie vole[114], horse[26], bovine[31], rat[29]and buffalo[32]with expression of lineage specific for endoderm, mesoderm and ectoderm (Table 1).Pluripotency is one of the defining features of iPS cells.Perhaps the most definitive test of pluripotency is the blastocyst complementation assay.The contribution of iPS cells to the resulting chimeras has been assessed to determine the differentiation capacity and germline contribution.True pluripotent murine iPS cells were generated using PB[115]and SB[21].To the best of our knowledge, there is no report on the successful transposon-derived iPS cellmediated germline contribution in large domestic animals.

Table 1 Differentiation potential of transposon-mediated induced pluripotent stem cells
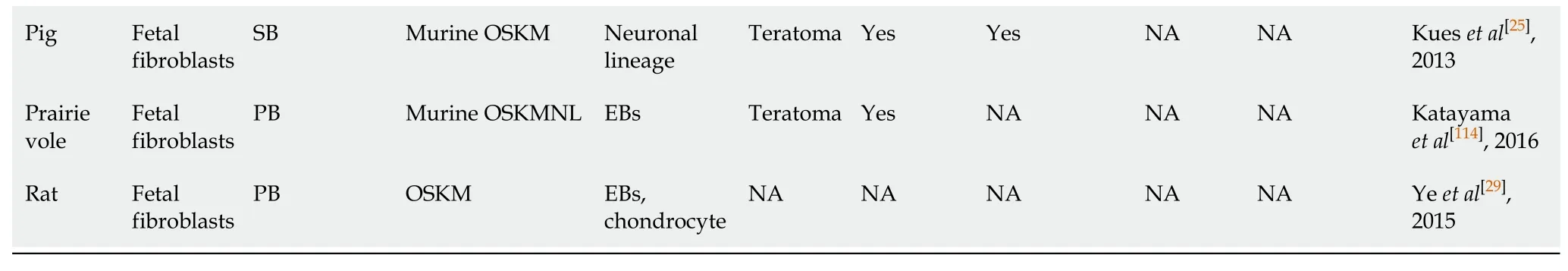
O: Oct4; S: Sox2; K: klf4; M: cMyc; N: Nanog; L: Lin28; EBs: Embryoid bodies; NA: Not applicable; PS: PiggyBac; SB: Sleeping beauty; AFP: α-Fetoprotein; SMA: Smooth muscle actin.

Figure 3 Timeline of transposon-mediated cellular reprogramming of porcine somatic cells to induced pluripotent stem cells (A), change in the morphology of somatic cells in the culture after transposition (B, unpublished own data), timeline of virus mediated cellular reprogramming of somatic cells to induced pluripotent stem cells (C).iPS: Induced pluripotent stem; MEF: Mouse embryonic fibroblast.
The iPS cells may be directed into the lineage of interest by supplementing various growth factors into the culture media.These growth factors or stimulating agents allow directed differentiation of iPS cells towards a particular cell lineage or cell type.The differentiated cells can be identified with the help of various markers, which are highly expressed in these cells.Very few markers are specific for one cell type, and as such, a panel of markers needs to be used in order to characterize the differentiation status.In this direction, EBs derived from SB-mediated mouse iPS cells were differentiated into cardiac cells with a beat frequency[21,23].Daviset al[20]observed that SB-mediated human iPS cells differentiated into EBs which contained hemoglobinized erythroid cells as well as spontaneously contracting cells, indicating that iPS cells could be differentiated into hematopoietic cell types and cardiomyocytes.
EBs generated from PB-mediated rat iPS cells showed numerous Alcian blue-stained regions, indicating the presence of acidic proteoglycans[29].These acidic proteoglycans were suggestive of cartilaginous tissue, which was further confirmed by the production of collagen II.Transgene-free human iPS cells derived from PB reprogramming were successfully differentiated into epidermal keratinocytes, which were found to be similar in morphological, functional, and molecular analysis of single-cell gene expression to normal human keratinocytes[116].The protocol for differentiation of human iPS cells into keratinocytes employed either retinoic acid or bone morphogenetic protein 4 (BMP4)[117].Igawaet al[116]used a modified protocol in which neither BMP4 nor retinoic acid were used.Around 5 weeks of initiation of differentiation, they reported obtaining keratinocyte-like cells.These cells were propagated through successive passaging at least five times in serum-free keratinocyte medium without feeder cells.Upon characterization, these cells were positive for K5/K14, suggesting successful differentiation of keratinocytes from human iPS cells, and they called these cells induced keratinocytes[116].These results indicate that iPS cell lines could be selected for therapeutic purposes.
Our group presented a novel approach for the differentiation of murine iPS cells derived through PB-mediated reprogramming into lentoid bodies[118].We established a co-culture system using human NTERA-2, a committed neuronal precursor cell line[119]and P19, a murine embryonic carcinoma cell line[120]to provide a suitable niche for differentiation of the iPSs into the ectodermal lineage.The developing lentoid bodies were identified by a lens lineage-specific reporter, but also showed changed light refraction in the bright-field view.The existing data support the notion that the specific cell type reporter approach is instrumental for the optimization, development and validation of differentiation protocols for murine iPS cells.We speculate that the gained knowledge can be translated to optimize the differentiation of lens cells from human iPS cells and thus to advance the progress of patient-specific lentoid bodies as a pipeline forin vitrodrug testing.It is likely that the specific cell type reporter approach is also adaptable forin vitrotracking of other cell lineages.
TRANSPOSON-BASED SYSTEMS FOR CELLULAR THERAPY
Cell-based therapy aims to treat diseases which cannot be addressed adequately by existing pharmaceutical interventions.The technology utilizes the cells with the ability to differentiate into specific lineages that are subsequently administered to a patient for therapeutic treatment.For this purpose, stem cells are considered ideal to restore tissue repair, or to replenish cells in the background of a genetic disease.The iPS cells can be expanded indefinitely and they are capable of differentiating in all the derivatives of the three germ layers.The generation of iPS cells is without the ethical stigma associated with ES cells, and iPS cells are able to result in personalized stem cells created from patient-specific cells.Although viral vectors are one of the most used methods for cellular reprogramming, their inherent limitations do not favor their clinical application due to hurdles in large-scale vector production and require careful biosafety characterization, which majorly impacts the costs of clinical-grade production of reprogrammed cells.
In recent years, non-viral DNA transposon based-systems have emerged as a potential tool to overcome some of the above-mentioned limitations.In transposonmediated genetic manipulation, gene(s) of interest such as therapeutic gene rendering stable phenotypic correction, can be introduced and the resulting stem cells can be expandedin vitroand then subjected to differentiation into particular cell lineages according to the therapeutic need.The iPS cells generated through transposonmediated cellular reprogramming are capable of differentiation into EBsin vitroand readily form teratomasin vivo.Teratoma formation confirmed that the reprogrammed iPS cells had the developmental potential to produce tissues of all three primary germ layers,i.e., ectoderm, mesoderm and endoderm[23,27,28,30,31].However, the gold standard of the iPS cells pluripotency is determined by their ability to form germline-competent chimeras.Woltjenet al[16]demonstrated the formation of murine chimeras from transposon-reprogrammed iPS cells.However, most of the currently used transposonmediated iPS cell lines carry constructs driven by a strong promoter, which constitutively promotes the reprogramming factors that will prevent the contribution to a normal ontogenesis[25,26,30].Thus, the transposon-mediated iPS cell lines in several species have not yet been tested for their capability to generate chimera and mediate germline transmission.The recent progress achieved in the area of integrationdeficient, but excision-competent transposase variants[61]will further simplify the transposon removal after complete reprogramming and the achievement of autonomous stemness.
Several advantages of transposon systems have encouraged investigators to carry out a clinical trial for the treatment of B-cell malignancies using SB-modified T-cell therapy[121].The results published in 2016 showed that the use of SB-modified chimeric antigen receptor (CAR) T-cells is safe when infused after allogeneic or autologous hematopoietic stem cell transplantation as an adjuvant therapy.Modified cells survived for an average of 51 or 201 d in the allogeneic or autologous setting, respectively, and patients showed progression-free survival rates that were improved when compared to historical data[122].Thereafter, iPS cell-based clinical trials have been initiated to treat Parkinson’s disease, heart disease and macular degeneration, highlighting the rapid progress that continues to be made in this area[123,124].To treat Duchenne muscular dystrophy, Filaretoet al[125]showed that SB-mediated ectopic expression of micro-utrophin in dystrophic iPS-derived skeletal muscle progenitors restored the muscle pathology by contributing to dystrophin–glycoprotein complex formation, which resulted in improved muscle contraction strength.PB-mediated expression of drug-inducibleMYOD1gene in human iPS cells lead to more efficient differentiation into myocytes[102].Similarly, SB-mediated overexpression of PAX3 in iPS cells induced differentiation into MYOD positive myogenic progenitors and produced multinucleated myofibers[126].Transposon-mediated iPS cells derived from patients suffering from either sickle cell disease caused by a β-globin gene mutation or Huntington’s disease caused by trinucleotide repeat expansions in the Huntingtin gene were successfully used for gene editing[127-129].The most commonly used transposons PB and SB were successfully used to generate human iPS cells from patient-derived cells with a disease-causing genetic background[16,22,130].These studies indicated that transposons are capable of introducing functional gene copies in patient-derived iPS cells containing defective genes.Recent evidence showed that transposon-mediated gene transfer was demonstrated in several types of cells such as ES cells, iPS cells, CD34+ hematopoietic stem cells or myoblasts[131].
Transposon-based gene delivery could also be used in combination with designer nucleases in iPS cells to correct gene defects.Yusa[132]reported that the endonucleasebased gene targeting efficiency increased using the PB transposon and it occurred due to the possibility of seamless removal of the drug marker enabled by re-transfection of the transposase.More recently, a transposon system was used in combination withCRISPR/Cas9for the generation of iPS cells from Huntington disease patients to correct mutations in the Huntingtin gene and corrected cells were then differentiated successfully into excitable, synaptically active forebrain neurons[129].Similarly, Wanget al[94]demonstrated that PB in combination with CRISPR/Cas9 for genome editing in iPS cells, in which the transposon deliveredCas9gene followed by delivery of sgRNA caused modification.Subsequent transient transposase expression of inducible Cas9 cassette was removed and yielded genome-edited iPS cells with seamless transgene removal.
The treatment of several human diseases often involves genetic manipulation of iPS cells prior to transplantation, which may further threaten their genomic stability.Overall, genomic aberrations can affect differentiation capability, identity and tumorigenicity of iPS cells.In the promising era of iPS cell research and therapy, the genomic stability of iPS cells and their safety, efficiency, and specificity remains one of the highest concerns prior to clinical translation[133].Hence preclinical trials in mice and other animal models are necessary in the future to confirm thein vivotherapeutic potential of reprogrammed cells.Challenges for reprogrammed cells are that they not only contain thein vivodelivery and dosage, but also their stability and potential offtarget effects[4].These challenges are currently hindering the progress to translate this potentially promising approach to clinical applications, but they appear to be solvable due to rapidly evolving advances in cellular reprogramming.
POTENTIAL RISKS OF TRANSPOSON-MEDIATED CELLULAR REPROGRAMMING AND THEIR SOLUTIONS
The use of SB systems appears to be safe in human cells with respect to off-target effects, as they originate from fish genomes, and the mammalian genome does not contain sufficient transposons to allow them to be cleavage by the transposase[50,73].Hence, the SB transposon exhibits the least deviation in genome-wide distribution and no apparent bias was observed for either the heterochromatic or euchromatic region and weak correlation with transcriptional status of targeted genes was detected[134].In addition, the ITRs region have negligible promoter/enhancer activity, and therefore they are unable to initiate transcription of genes that flank the integration site[135].This system is highly efficient in transfecting even those cell types which are hard to transfect.On the other hand, PB systems have a wide target site that favor integration into genes and near chromatin marks characteristic of active transcription units[73,134,136,137].These observations indicate that transposons (SB and PB) might be safe for therapeutic gene delivery in clinical trials.
After delivery of the transposon system, the transposition may undergo multiple rounds of remobilization[138,139], which should be minimized by carefully controlling the transposase dose[136].In mouse embryonic stem cells approximately 95% of genomic transposon excision was reported to be precise and 5% of the transpositions showed genomic alterations[138].It was also observed that frequent transposition into unknown sites could result in micro-deletions, footprint mutations as well as chromosomal rearrangements in the genome, which makes it labor intensive to identify integrationfree iPS cells with intact genomes[138,140].As a consequence, the transposase expression window should be tightly controlled to achieve traceless excision without inducing any genomic alterations and cytotoxicity[141].
Due to its non-viral nature and integration capacity, some of the transposon systems were adapted for use in gene therapy practices.To achieve efficient and safe use, the transposon systems were split into two plasmids, one containing the sequence encoding the transposase enzyme and the other comprising an expression cassette flanked by ITRs.However, in spite of these advantages, DNA transposon based vectors are essentially gene-inserting tools that still need assistance for efficient cellular uptake.Therefore, its activity depends on cell type, transfection method, and plasmid size.Moreover, it is important to note that these vectors have been largely used in the preclinical setting, and clinical trials are in progress to evaluate their efficacy, safety and presumed advantages.
Transposon-based gene transfer followed by cellular reprogramming might be associated with the important risk factor of genotoxicity.The genotoxicity could be induced either by interaction of the transposase with endogenous DNA sequences, or the genome-wide insertion profile of the transposon vector.To increase the efficacy and safety of cellular reprogramming, many efforts have been made to obtain potential molecules that can improve reprogramming efficiency or replace some of the vital transcription factors[142].In this direction, various small molecules such as histone deacetylase inhibitors, DNA methyltransferase inhibitors, methylases, and demethylase inhibitors, Rho-associated protein kinase, and Wnt pathway regulators have been recognized to be effective in inducing reprogramming of terminally differentiated cells[143-146].Huangfuet al[147]showed that valproic acid, a histone deacetylase inhibitor, increased the efficiency of transcription factor-mediated cellular reprogramming from 0.50% to 11.8%, indicating chromatin modification is one of the major rate-determining steps during cellular reprogramming.In addition to these, other molecules have also been tested to improve cellular reprogramming efficiency, including RepSOX2, E-616452 (2-[3-(6-methyl-2-pyridinyl)-1H-pyrazol-4-yl]-1,5-naphthyridine), and OAC1 (Oct4-activating compound 1), which facilitate the mesenchymal-epithelial transition (MET), and activate the stemness-associated promoter regions of mature fibroblasts[148,149].Nowadays, the use of these small molecules is more trustworthy for introducing transcription factors into cells, but it remains a challenge to break through the efficiency threshold due to inadequate gene delivery and limitations in cellular uptake[150].
As compared to integration of retrovirus[151]and lentivirus[152], the integration profile of PB[137]and SB are safe, and are currently being tested for several clinical trials of T cell immunotherapy.Furthermore, to exclude the possibility of remobilization, the transposase could be transfected in the form of RNA, which seems to be less toxic to the cells[153].
CONCLUSION
Recently, transposon systems have been developed as attractive tools for somatic cell reprogramming, which has significant potential in speeding up patient-specific cell based therapies, as they can overcome some of the limitations of viral-based reprogramming technologies[125,154].Furthermore, transposon systems have unique features for excising the exogenous reprogramming cassette from the iPS genome through re-expression of the transposase.Transposon systems eventually gives rise to “transgene-free” iPS cells, which is valuable in minimizing the risk of reactivation of reprogramming factors with oncogenic potential[94].In addition to gene delivery, gene correction can also be achieved with a combination of transposons and designer endonucleases including ZFN, TALEN or CRISPR/Cas9.The introduction of a sitespecific DNA double-strand break by endonuclease activity allows homologous recombination at target genes, followed by traceless removal of selectable gene cassettes by the transposase.This strategy has been used in SCD patient-derived iPS cells without any detectable off-target activity and undesirable chromosomal alterations[127].More recently, the practice of mRNA encoding transposases to prevent continued mobilization of transposons and modification of ITRs, and the generation of hyperactive and codon-optimized transposase variants enhanced the overall transposition efficiencies[88].This broadens the spectrum of possible therapeutic alternatives for gene therapy in particular, and gene correction in iPS cells.A number of preclinical studies performed as disease models that simulate the cognate human disorders have highlighted the potential of transposons for gene therapy[154].Thus, iPS cell biology will continue to play a major role not only in the advancement of medical sciences, but also in improving the understanding of basic sciences.Looking forward, the continued advancement and refinement of transposon based-technologies and the steps toward their clinical translation will likely herald an exciting era in gene therapy.
 World Journal of Stem Cells2020年7期
World Journal of Stem Cells2020年7期
- World Journal of Stem Cells的其它文章
- Approaches to promoting bone marrow mesenchymal stem cell osteogenesis on orthopedic implant surface
- Photodynamic therapy regulates fate of cancer stem cells through reactive oxygen species
- Decellularized adipose matrix provides an inductive microenvironment for stem cells in tissue regeneration
- Vitamin D and calcium signaling in epidermal stem cells and their regeneration
- Adipose-derived stem cell therapy shows promising results for secondary lymphedema
- Involvement of glycated albumin in adipose-derived-stem cell-mediated interleukin 17 secreting T helper cell activation
