Early detection of non-small cell lung cancer in liquid biopsies by ultrasensitive protease activity analysis
Dinusha N. Udukala, Sebastian O. Wendel, Hongwang Wang, Asanka S. Yapa, Obdulia Covarrubias-Zambrano, Katharine Janik, Gary Gadbury, Deryl L. Troyer, Stefan H. BossmannDepartment of Chemistry, Kansas State University, Manhattan, KS 66506, USA.
2Department of Statistics, Kansas State University, Manhattan, KS 66506, USA.
3Department of Anatomy & Physiology, Kansas State University, Manhattan, KS 66506, USA.
#Authors contributed equally.
Abstract Aim: A significant fraction of mortalities from non-small cell lung cancer could be prevented, if the cancer would be diagnosed earlier. Nanobiosensors for the ultrasensitive detection of active proteases in serum were designed to detect a significant protease activity signature of non-small cell lung cancer (stage I and higher).Methods: We determined the activity of nine protease biomarkers in the sera of non-small cell lung cancer patients and compared them with the protease activities of a control group of healthy human subjects using optical nanobiosensors. They consist of a central Fe/Fe3O4 core/shell nanoparticle with an attached Fluorescence resonance energy transfer-pair [tetrakis-carboxyphenyl porphyrin (TCPP) and cyanine 5.5]. TCPP is attached to the central nanoparticle via a protease-cleavable tether, whereas cyanine 5.5 is tethered permanently to the dopamine-layer surrounding the nanoparticle. Results: Based on the activity pattern of urokinase plasminogen activator, matrix metalloproteinases 1, 2, 3, 7, 9, and 13, and cathepsins B and L as well, non-small cell lung cancer could be detected at stage I by means of a liquid biopsy. Conclusion: This feasibility study, comprising 33 non-small cell lung cancer patients and 20 apparently healthy subjects, clearly demonstrated the feasibility of minimally invasive early diagnosis of non-small cell lung cancer, starting with stage I.
Keywords: Non-small cell lung cancer, lung cancer diagnosis, protease activity, liquid biopsies, iron/iron oxide nanoparticles, significance table, multivariate model
INTRODUCTION
Lung cancer is the second most common cancer in men and women. For 2020, a total of 228,820 new cases are estimated. For both genders, lung cancer is on top of the cancer mortality ranking in the US, and 135,720 cancer mortalities are anticipated this year[1]. Approximately 13% of all lung cancers are small cell lung cancers (SCLC), and 84% are non‐small cell lung cancers (NSCLC). Here, we focus on the detection of NSCLC by means of optical nanobiosensors capable of determining the activity of signature proteases in serum. In Figure 1, the 5‐year survival of NSCLC patients as a function of cancer stage at the time of lung cancer diagnosis is shown. The staging system used for NSCLC in this report is the American Joint Committee on Cancer (AJCC) TNM system, which is based on the size and extent of the main tumor (T), the spread to nearby lymph nodes (N), and thzhegnwkais e metastasis to distant sites (M)[2]. It is clearly discernible that the 5‐year survival rate significantly decreases if the cancer is diagnosed late. This implies that early detection of cancer saves lives. On the basis of the 5‐year survival statistics of NSCLC, we conservatively estimate that about 30 percent of mortalities could be prevented if NSCLC were routinely detected at stage 1 (combined T1a, T1b, and T1c) instead at stages 2 or 3, when it is currently diagnosed[3]. The optical nanobiosensors developed in the Bossmann group feature sub‐femtomolar limits of detection, thereby permitting the diagnosis of NSCLC by means of a liquid biopsy utilizing the serum of cancer patients[4-7].
Liquid biopsies
In 2000, Veridex introduced the first commercially available liquid biopsy assay, the CELLSEARCH®CTCtest[8]. In 2016, the Food and Drug Administration (FDA) approved the cobas® epidermal growth factor receptor (EGFR) Mutation Test for determination of the EGFR gene mutation in blood from lung cancer patients[9]. Commercially available liquid biopsies consist in the detection of circulating tumor cells (CTCs), tumor‐derived exosomes, circulating cell‐free DNA (cfDNA), microRNA (miRNA), signaling proteins and metabolic enzymes (proteases and kinases)[10]. Virtually all companies in this field, among them Personal Genome Diagnostics[11], Genomic Health[12], Myriad Genetics[13], Guardant Health[14]and Pathway Genomics[15]rely on PCR to detect genetic mutations, and various RNAs that are overexpressed in tumors[11-15]. CancerSEEK evaluated the plasma levels of 8 cancer‐indicating proteins and the presence of mutations in 2,001 genomic positions for detecting 8 solid tumor types, among them pancreatic cancer with an overall percentage of cancer detection of 62% at > 99% specificity[16]. DETECT‐A (detecting cancers earlier through elective mutation‐based blood collection and testing) was able to detect 26 cancers by means of a liquid biopsy (nine lung cancers, six ovarian cancers and two colorectal cancers). Seventeen (65%) of the 26 cancers were localized or regional, including five patients with stage I tumors[17]. The GRAIL technology looks at differences in DNA methylation between cancerous and healthy cells. It is capable of detecting > 50 different cancers from one liquid biopsy and has a very low false positive rate. Regardless of the cancer detection technology used, it is of vital importance to detect cancers at AJCC stage I or earlier to maximize cancer survival[17]. GRAIL is promising early detection capability, because changes in DNA methylation occur early in cancer[18,19]. As for all approaches, including the optical nanobiosensors for protease activity profiling discussed here, future comparative clinical trials will permit a true comparison of genomic vs. other methods of early cancer detection.
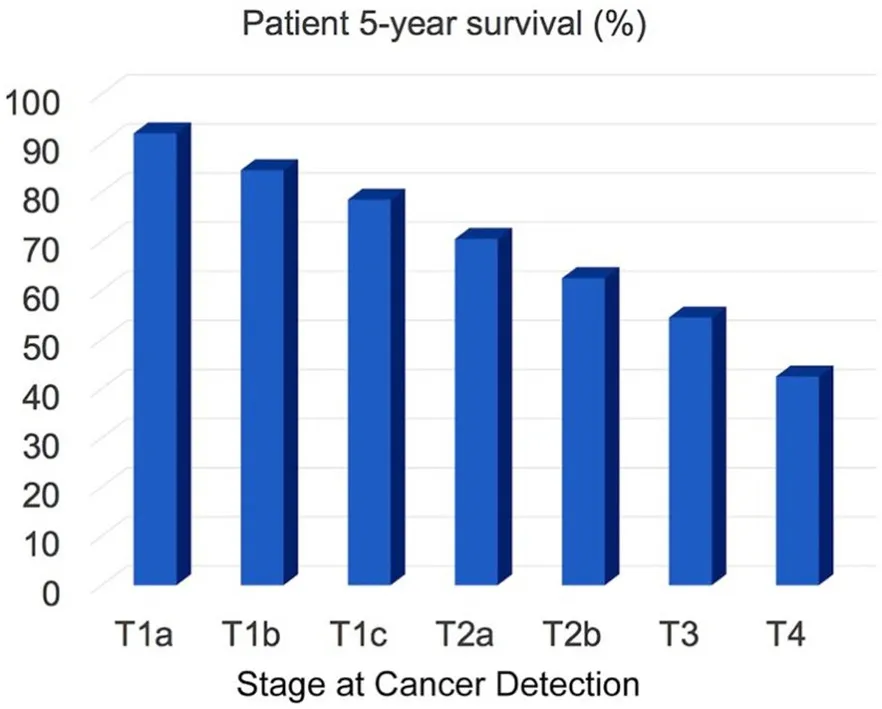
Figure 1. Patient 5-year survival (in percent) as a function of cancer stage at the time of cancer diagnosis. Data from references[1-3]
Optical nanobiosensors for protease detection
It is established that virtually all solid tumors are characterized by dysfunctional protease expression patterns[20-22]. These deviations from the proteasome of healthy cells have been successfully used by the authors for the early detection of breast[5]and pancreatic[7]cancer. In cancer, numerous proteases, such as matrix metalloproteinases (MMPs)[23], cathepsins[24-26], and urokinase plasminogen activator (uPA)[20,22,25]are either over‐ or underexpressed, when compared to healthy cells. This enables the detection of solid tumors in liquid biopsies through the simultaneous detection of several proteases in serum[5,7].
The Bossmann group has continuously developed their patented technology for ultra‐sensitive protease detection since 2007[27-29]. These fluorescence‐based optical nanobiosensors are composed of water‐dispersible dopamine‐coated Fe/Fe3O4core/shell nanoparticles and an attached Fluorescence resonance energy transfer pair[30][tetrakis (4‐carboxyphenyl) porphyrin (TCPP) and cyanine 5.5]. Both the central nanoparticle and cyanine 5.5 are able to quench photoexcited TCPP, which is attached via a protease‐cleavable consensus sequence[4]. The consensus sequences have been optimized to show only minimal cross‐sensitivity between the proteases used[5][Figure 2].
Selection of protease biomarkers
About two percent of the human genome encodes proteases, 553 in total[31]. For detecting NSCLC, we selected the following proteases: MMPs 1, 2, 3, 7, 9 and 13, uPA and cathepsins B and L. This selection is identical to that in a previous study, in which breast cancer at stage 1 could be detected[5]. Evidently, we have ascertained that this selected group of proteases is indeed implicated in NSCLC[20,22,23,25,26]. The advantage of using the same panel of proteases for detecting breast cancer and non‐small cell lung cancer is that we can compare the resulting “significance tables” and begin to find answers to the question whether protease expression pattern of different types of solid tumors are similar or significantly different. In Table 1, the consensus sequences for the nine proteases used are summarized. Note that the cross‐reactivity of these consensus sequences is reported in reference[5]. With the exception of MMP9 and MMP2, which show about 20%, only minor cross reactivities (< 5 rel. %) were observed.

Figure 2. Fe/Fe3O4-based nanobiosensor for matrix metalloproteinase 1 detection. Consensus sequence GAGVPMS-MRGGAG. TCPP: tetrakis-carboxyphenyl-porphyrin. Scheme 1. A: Design principles of a nanobiosensor for protease detection. Consensus sequence: GAGVPMS-MRGGAG (MMP1); The average distance between fluorophore TCPP (tetrakis-carboxyphenyl-porphyrin) and Fe/Fe3O4 nanoparticle and FRET-acceptor cyanine 5.5 increases upon cleavage of the oligopeptide tether by a suitable protease, leading to an increase in fluorescence; B: TEM and HRTEM of dopamine-coated Fe/Fe3O4 core/shell nanoparticles; C: Typical emission spectra occurring from the nanosensor for MMP 13 after 1 h of incubation at 37 oC (λexc = 421 nm). low: buffer; middle:nanosensor; high: nanosensor after incubation with MMP13[4].
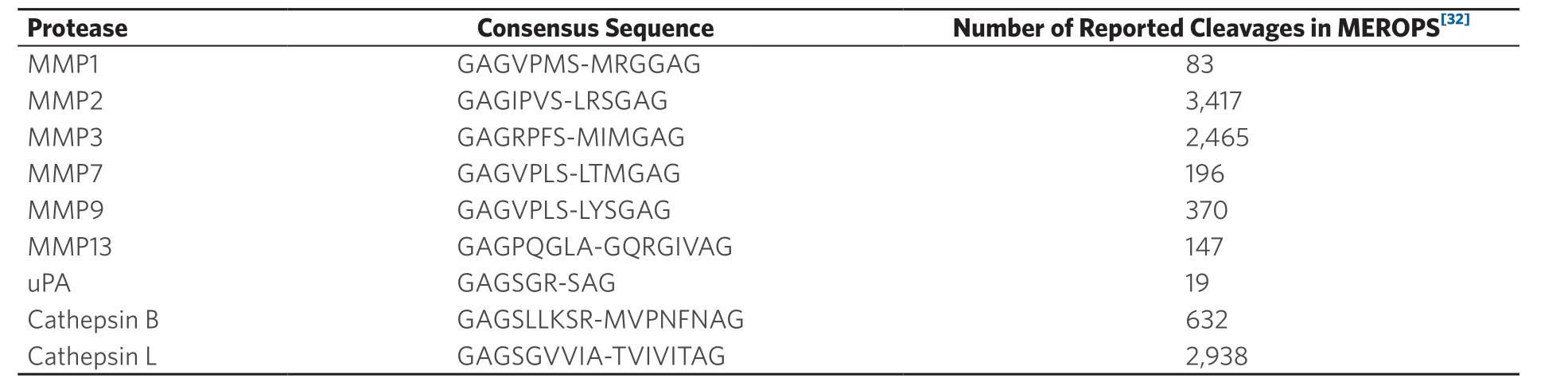
Table 1. Consensus sequences in single-letter code for nine proteases that have been identified as cancer biomarkers. Essential amino acids of the consensus sequences are in bold[32]
METHODS
Serum samples
We obtained 53 de‐identified human serum samples (at ‐80oC) from Dr. Tracy Chapman, Southeastern Nebraska Cancer Center (SNCC). All subjects involved in this study were Caucasian. Twenty were apparently healthy volunteers (ages 26 to 68), and 33 were NSCLC patients {9 patients were diagnosed with stage 1 [T1a (3), T1b (2) and T1c (4)], 12 patients were stage 2 [T2a (5) and T3a (7)], and 12 patients were stage 3 (T3)}. The NSCLC patients ranged from 35 to 70 in age. In a previous study, we established that the protease expression pattern of healthy and (breast) cancer patients was not significantly affected by gender. The underlying paradigm of this study was that also in lung cancer, there are no gender differences in protease expression patterns of cancer and healthy human subjects.
Statistical analysis
Bar graphs and box plots to illustrate data ranges and P‐values[33]were created using the R software environment for statistical computing[34,35]. To determine possible differences between cancer patients and healthy control groups for each protease nanobiosensor, P‐values were derived by using an unpaired and two‐sided Student t‐test with Welch modification. P‐value (calculated probability) is the probability of finding the observed results if the null hypothesis of a study question is true. The null hypothesis assumes that there is no difference between a selected property of a group of study subjects vs. a control group[36].
Nanobiosensor synthesis
Detailed descriptions of the syntheses of nanobiosensors and the required components (dopamine Fe/Fe3O4nanoparticles, oligopeptides, TCPP, and cyanine 5.5) have been reported in our previous studies[15,16,18,19]. Briefly, the nanobiosensors were assembled from dopamine coated Fe/Fe3O4nanoparticles, cyanine 5.5, and peptide sequences that were linked to TCPP on resin[15]. Thirty‐five TCPP and 50 cyanine 5.5 dye molecules were attached on average to each Fe/Fe3O4core/shell nanoparticle, on the basis of statistical modelling[37,38]. To achieve this synthesis, a solution was prepared by completely dissolving 64 mg of TCPP‐linked peptide sequence, 37 mg cyanine 5.5, 45 mg EDC and 45 mg DMAP in 30 mL of anhydrous DMF. In a separate vial, 450 mg of dopamine‐coated Fe/Fe3O4nanoparticles were dispersed in 10 mL of anhydrous DMF by sonicating for 20 min. Both solutions were then mixed, sonicated for 10 min, and incubated overnight on a shaker at room temperature. After overnight incubation, the resulting nanobiosensor was collected via centrifugation (5 min at 10,000 rpm), washed with DMF to remove excess dye and unbound components, followed by five washes with cold ether (‐10oC). After each washing step, the nanobiosensor was collected via centrifugation. The nanobiosensors were then collected and dried with argon. The nanobiosensors could be stored for more than 1 year at ‐20oC under argon.

Standard procedure of preparing protease assays
For each protease, 3.0 mg of nanoplatform were dispersed in 3.0 mL of PBS (phosphate‐buffered saline, Aldrich). The suspension was sonicated for 10 min, and 3.0 mL of PBS‐dextran (10 mg dextran in 1.0 mL of PBS) were mixed with 75 μL of the nanoplatform suspension (3.0 mg in 3.0 mL of PBS, see above) and 30 μL of serum. The suspensions were incubated at 37oC for 60 min, followed by the recording of a fluorescence spectrum at 25oC using a Fluoromax2 spectrometer (λem= 421 nm, λex= 620‐680 nm).
RESULTS
The major advantages of the fluorescent Fe/Fe3O4core/shell nanoparticle‐based nanobiosensors are their sub‐femtomolar limits of detection, their large ranges extending over at least eight orders of magnitude, and their reliably small experimental errors of ± 3 percent. The calibration of the optical nanobiosensors with commercially available proteases, as well as a modest serum effect on their calibration curves are discussed in detail in an earlier study[5]. We also established that the statistical analysis of the protease activity in the serum of cancer patients vs. a healthy control group of volunteers can be performed by using the integrated fluorescence signal of the nanobiosensors[5]. Our cancer detection method by means of liquid biopsies relies on the paradigm that the extensive protease network of human cells is dysfunctional in cancer[22]. Therefore, unique protease signatures can be established for virtually any solid tumor. We furthermore hypothesized that tumors derived from different cell types should exhibit different protease activity patterns and, therefore, unique protease signatures.
The activities of nine selected proteases (cathepsins B and L, uPA, and MMP1, 2, 3, 7, 9, and 13) in the serum of 33 NSCLC patients and 20 healthy volunteers were determined following the procedure described in the experimental section. In short, each nanobiosenor designed for measuring the activity of one protease and a small serum sample (30 μL) were incubated in dextran‐containing PBS buffer for 1 h at 37oC. The dextran was added to prevent the coagulation of the nanobiosensor suspension during the time required for protease sensing. After 1 h, the resulting fluorescence of the suspension was measured, which indicates the protease activity in serum. A series of boxplots and bar graphs [Figures 3‐7] show the data range for each protease that correlates to each cancer stage, as well as the protease activity range of healthy patients.
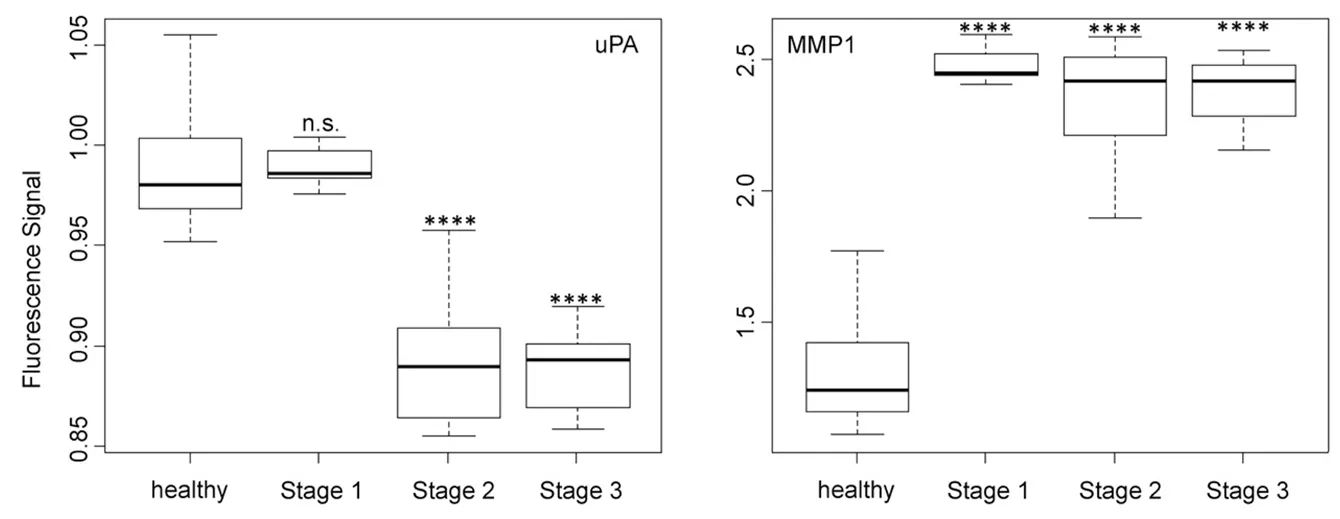
Figure 4. Box plots (indicating the observed data range) for urokinase plasminogen activator and matrix metalloproteinase 1. The groups are apparently healthy control group (n = 20), non-small cell lung cancer stage 1 (n = 9), non-small cell lung cancer stage 2 (n = 12), and non-small cell lung cancer stage 3 (n = 12). ⋆P < 0.05; ⋆⋆P < 0.01; ⋆⋆⋆P < 0.001; ⋆⋆⋆⋆P < 0.0001; n.s.: not significant. All biospecimens were obtained from the South Eastern Nebraska Cancer Center. Non-small cell lung cancers had been staged according to the American Joint Committee on Cancer TNM staging system[2]
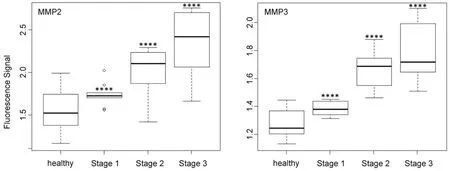
Figure 5. Box plots (indicating the observed data range) for matrix metalloproteinase MMP2 and MMP3. The groups are apparently healthy control group (n = 20), non-small cell lung cancer stage 1 (n = 9), non-small cell lung cancer stage 2 (n = 12), and non-small cell lung cancer stage 3 (n = 12). ⋆P < 0.05; ⋆⋆P < 0.01; ⋆⋆⋆P < 0.001; ⋆⋆⋆⋆P < 0.0001. All biospecimens were obtained from the South Eastern Nebraska Cancer Center. Non-small cell lung cancers had been staged according to the American Joint Committee on Cancer TNM staging system[2]
As shown in Figure 3, cathepsin B was expressed equally by the control group and the group of stage 1 NSCLC patients, whereas the distinct upregulation of cathepsin B activity was observed for stage 2 and 3 patients. Although cathepsin B was unsuited for detecting (sub)stage 1 NSCLC, it provides data for cancer staging via liquid biopsy. In opposite to cathepsin B, cathepsin L showed a steady increase in activity with increasing stage of NSCLC. This behavior is well suited for staging and also permits the detection of stage 1.
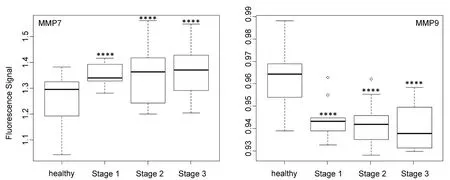
Figure 6. Box plots (indicating the observed data range) for matrix metalloproteinase MMP7 and MMP9. The groups are apparently healthy control group (n = 20), non-small cell lung cancer stage 1 (n = 9), non-small cell lung cancer stage 2 (n = 12), and non-small cell lung cancer stage 3 (n = 12). ⋆P < 0.05; ⋆⋆P < 0.01; ⋆⋆⋆P < 0.001; ⋆⋆⋆⋆P < 0.0001. All biospecimens were obtained from the South Eastern Nebraska Cancer Center. Non-small cell lung cancers had been staged according to the American Joint Committee on Cancer TNM staging system[2]
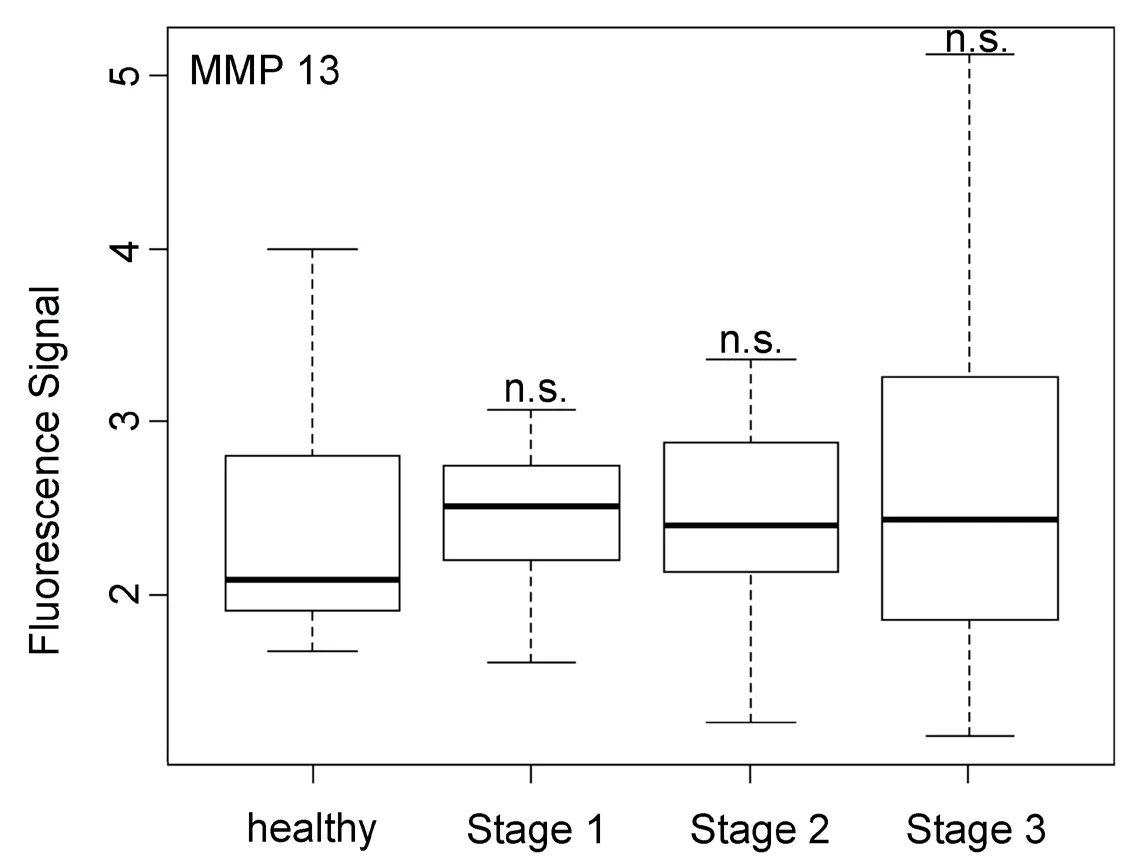
Figure 7. Box plots (indicating the observed data range) for matrix metalloproteinase 13. The groups are apparently healthy control group (n = 20), non-small cell lung cancer stage 1 (n = 9), non-small cell lung cancer stage 2 (n = 12), and non-small cell lung cancer stage 3 (n = 12). ⋆P < 0.05; ⋆⋆P < 0.01; ⋆⋆⋆P < 0.001; ⋆⋆⋆⋆P < 0.0001; n.s.: not significant. All biospecimens were obtained from the South Eastern Nebraska Cancer Center. Non-small cell lung cancers had been staged according to the American Joint Committee on Cancer TNM staging system[2]
The activity of uPA [Figure 4], which is an originally misnamed protease, depended on the NSCLC staging in a very similar manner as cathepsin B. It is noteworthy that the optical nanobiosensors for uPA and MMP9 decreased with increasing protease activities, whereas all other protease nanobiosensors increased in fluorescence. Similar to cathepsin B, uPA was well suited for NSCLC staging but not for early cancer detection. A very different type of dependence of nanobiosensor fluorescence on NSCLC staging was observed for MMP1 activity, which was significantly increased starting at stage 1! However, it did not change significantly when progressing to stages 2 and 3. This finding makes MMP1 our best biomarker for very early diagnosis of NSCLC.
In contrast to MMP1 and very similar to cathepsin L, the protease activities of MMP2, MMP3, MMP7, and MMP9 in the serum of NSCLC patients increased steadily with escalating cancer stage [Figures 5 and 6]. However, it should be noted that for all three MMPs, their expression levels in the sera of NSCLC patients at stage 1 and the healthy control group were statistically different.
In opposite to all other protease activities studied here in the serum of NSCLC patients, MMP13 was found to be unsuited for both early cancer diagnosis and NSCLC staging. As shown in Figure 7, MMP13 activity was basically the same for healthy subjects and NSCLC patients of all three investigated stages. This finding was unexpected, because MMP13 is known to be involved in endothelial‐mesenchymal transition[39].
DISCUSSION
The sensitivity of the Fe/Fe3O4nanoparticle‐based fluorescence nanobiosensors permits the accurate measurement[5]of the activities of nine signature proteases in serum samples (30 μL) obtained from NSCLC patients. This technology permits the rapid and inexpensive detection of NSCLC at stage 1 by means of a simple liquid biopsy. We estimate costs of approx. $20 for measuring the activity of the 8 required proteases in serum. Six proteases permit the detection of NSCLC at stage 1. MMP1 is the best candidate for the detection of NSCLC, due to the large increase in activity of 3.16 × 10‐9mol/L of stage 1 patients compared to 1.35 × 10‐15mol/L the control group. Principally, a protease‐based liquid biopsy for NCSLS has the potential of significantly reducing lung cancer mortality, because lung cancer treatment would be more successful when the cancer would be detected at stage 1 instead of stages 2 and 3, which is usually the case at present day. It should be noted that this technology works for the detection of virtually all solid tumors, of which many feature distinct protease signatures. When comparing the protease signatures of breast cancer[5]and NSCLC, there are similarities and differences: (1) for all investigated proteases, increased activities are detected for consecutively higher cancer stages, compared to the control group of healthy subjects; (2) Cathepsin B is more significant for breast cancer than for NSCLC; (3) on the other hand, MMP7 is far more sensitive in detecting NSCLC than breast cancer; and (4) MMP13 is an unsuitable biomarker for cancer detection in either case. Based on this comparison, it is our conclusion that the similarities between the protease expression pattern of NSCLS and breast cancer far outweigh the differences. Therefore, the panel of proteases should be expanded to detect characteristic differences, which could be used for the identification of the type (and the stage) of solid tumor by means of a liquid biopsy. The latter should be offered in regular intervals to all members of cancer risk groups.
The “Significance Table 2” summarizes our findings. It shows 95% confidence intervals and P‐values[33]calculated for the comparison of the members of individual NSCLC stages with the group of healthy volunteers. P‐values < 0.05 are considered significant and shown in green. Cathepsin L, and MMP1, 2, 3, 7, and 9 permit the detection of NSCLC at stage 1. The statistically non‐significant P‐values are marked in red.
In Table 3, the average activities for all nine investigated proteases are summarized. The calibration curves in the presence of human serum that are discussed in reference[5]were run again in parallel to the protease measurements described here. They were used to calculate the protease activities for NSCLC patients and the control group of healthy volunteers. To date, most of the protease measurements in cancer research and clinical diagnosis are performed by means of immunoassays[10]. Whereas the latter measure the total concentration of protease, our optical nanobiosensors determine only the fraction of active proteases. As discussed earlier, a complex protease network exists in human biology that has the ability to form activation cascades[20,31]. Furthermore, protease zymogens can act as signaling peptides, depending on their glycosylation pattern[40]. Comparing the concentrations of active and inactive proteases in cancer and numerous other diseases may offer an unprecedented insight into the human proteasome and also provide diagnostic opportunities. A combination of MMP1 and cathepsin B was chosen for a multivariate model. MMP1 achieved a good separation between the healthy group and stage 1, while cathepsin B increased the separation between the later stages, a quality that MMP1 lacked. The parameter’s values were linearized (log) and this model’s coefficients were highly significant. The R2‐value improved to 0.91. The graph shows how the data groupings shift upward (MMP1 influence) and to the right (cathepsin B influence) with increasing cancer stages [Figure 8]. This would allow for a precise detection of stage 1 NSCLC.

Table 2. Significance table: protease expression pattern of non-small cell lung cancer at stages 1, 2, and 3

Table 3. Activities of cathepsins, MMPs and uPA in serum as a function of stage
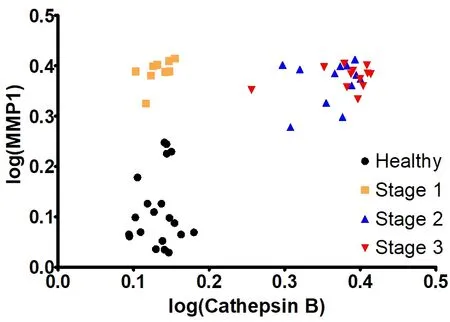
Figure 8. Multivariate model: statistical analysis: P-value of matrix metalloproteinase (MMP)1 slope: 4.34e-13 MMP1 slope: 0.90623; P-value of cathepsin B slope: 9.11e-15l and cathepsin B slope: 1.13514; R2 value: 0.9181
DECLARATIONS
Acknowledgments
The authors thank Dr. Tracy Chapman, Southeastern Nebraska Cancer Center (SNCC) for the biospecimens used in this study.
Authors’ contributions
Synthesis and characterization of the optical nanobiosensors: Udukala DN, Wendel SO, Wang H, Yapa AS
Calibration of the optical nanobiosensors: Udukala DN
Liquid biopsies: Udukala DN, Covarrubias‐Zambrano O
Data analysis: Udukala DN, Janik K
Statistical data analysis: Wendel SO, Covarrubias‐Zambrano O, Gadbury G
Design of the study reported here: Troyer DL, Bossmann SH
Writing of the manuscript: Covarrubias‐Zambrano O, Janik K, Bossmann SH
Availability of data and materials
The raw data will be made available upon request; please contact Dr. Stefan H. Bossmann.
Financial support and sponsorship
National Science Foundation, USA, grant (CBET 1804416).
Conflicts of interest
All authors declared that there are no conflicts of interest
Ethical approval and consent to participate
The biospecimens were collected by Dr. Tracy Chapman, Southeastern Nebraska Cancer Center (SNCC).
We received de‐identified patient information.
Consent for publication
The deidentified serum samples were purchased from the South Eastern Nebraska Cancer. The activities at Kansas State were classified as “non research” with respect to the biospecimens used.
Copyright
© The Author(s) 2020.
 Journal of Cancer Metastasis and Treatment2020年8期
Journal of Cancer Metastasis and Treatment2020年8期
- Journal of Cancer Metastasis and Treatment的其它文章
- The systemic hallmarks of cancer
- Targeting the JAK/STAT pathway in solid tumors
- Perspective on dietary isothiocyanates in the prevention, development and treatment of cancer
- Burden of chronic diseases among sarcoma survivors treated with anthracycline chemotherapy: results from an observational study
- Targeting epithelial-mesenchymal transition - an ongoing wild goose chase
