A PCR and RFLP-based molecular diagnostic algorithm for visceral leishmaniasis
Natalia Souza de Godoy, Manoel Sebastião da Costa Lima-Junior, José Angelo Lauletta Lindoso, Vera Lucia Pereira-Chioccola, Thelma Suely Okay, Lucia Maria Almeida Braz,5
1Laboratório de Parasitologia Médica, Departamento de Moléstias Infecciosas e Parasitárias do Hospital das Clinicas HCFMUSP, Faculdade de Medicina, Universidade de São Paulo, São Paulo, Brazil
2Laboratório de Imunopatologia e Biologia Molecular-Instituto Aggeu Magalhães/IAM-FIOCRUZ, Recife, Brazil
3Laboratório de Soroepidemiologia e Imunobiologia-FMUSP, São Paulo, Brazil
4Laboratório de Biologia Molecular de Parasitas e Fungos-Instituto Adolfo Lutz, São Paulo, Brazil
5Laboratório de Parasitologia-Instituto de Medicina Tropical de São Paulo-USP, São Paulo, Brazil
ABSTRACT
KEYWORDS: Leishmania infantum; Molecular diagnosis;Visceral leishmaniasis; PCR; RFLP
1.Introduction
According to the World Health Organization (WHO), 300 000 cases of visceral leishmaniasis (VL) are reported worldwide annually with 40 000-50 000 deaths[1].In the Americas, 90% of cases occur in Brazil, and Leishmania (L.) infantum (donovani complex) is the causative agent of VL[2].Accurate and rapid diagnosis of VL is necessary, because it can be lethal.Parasitologic and serologic methods (gold standard) are used for the laboratory diagnosis of VL;however, they have low sensitivity, especially the serologic methods in immunocompromised patients.Polymerase chain reaction (PCR)is more sensitive[3,4] and specific[3,5] than these routine methods[1,6].In addition, it is possible to identify the species of Leishmania using different targets and methodologies.In some situations, identification of Leishmania is crucial, especially for patients coinfected with Leishmania and HIV[2,7,8], because the dermotropic species (L.braziliensis and L.amazonensis) can cause visceral lesions[9-11].On the other hand, cutaneous and mucosal leishmaniasis can be caused by L.infantum[8,12] in both immunocompetent and immunosuppressed patients.Therefore, this wide range of clinical presentations make identification of the causative agent necessary,and molecular biology techniques can play an important role[5,9- 13].Identification of the Leishmania species is also important to identify the species circulating in a given area, especially in regions where different species are present, as occurring in Brazil.Using molecular biology, various sequences from both genomic and extrachromosomal regions have been exploited as targets of amplification by PCR[14], such as Leishmania kinetoplast DNA (kDNA) RV1/RV2 and Leishmania internal transcriber spacer DNA (ITS1 DNA)LITSR/L5.8S.PCR using kDNA primers presents high sensitivity,because kDNA is present in large numbers within the mitochondria of the parasite.ITS sequences are composed of highly conserved regions, allowing their use in PCR for diagnostic purposes, and they have polymorphic regions that can be used in restriction fragment length polymorphism (RFLP) assays to determine Leishmania species[15].In addition, bone marrow aspirate (BMA) and peripheral blood (PB) can be used as sources for L.infantum DNA research[6].Therefore, in order to diagnose VL and identify the Leishmania species causing VL, we aimed to determine an algorithm for the molecular diagnosis of VL using kDNA PCR (RV1/RV2) and ITS1 PCR (LITSR/L5.8S), complemented by ITS1 PCR RFLP, using PB or BMA from patients with suspected VL.
2.Materials and methods
2.1.Location of the study and ethical approval
After approval of the Commission for Research Projects Analysis(CAPPesq, process number 191.806/2013), the study was carried out between July 2014 and November 2016 in Instituto de Medicina Tropical da Universidade de São Paulo (Tropical Medicine Institute),São Paulo, Brazil.
2.2.Biological samples
The biological samples used in this study were obtained from seven groups as follows.GroupⅠ: 82 samples (PB and/or BMA)were obtained from patients attending the Hospital das Clínicas da Faculdade de Medicina da Universidade de São Paulo (HCFMUSP)and at the Universidade Federal do Mato Grosso do Sul (UFMS)with VL confirmed by clinical, epidemiologic, parasitologic, and/or serologic examinations performed by the immunochromatographic rapid test (IrK39).GroupⅡ: 16 samples from patients under treatment for VL, confirmed by clinical, epidemiologic, parasitologic,and/or serologic examination (data extracted from medical records).GroupⅢ: 14 BMA samples from dogs with canine visceral leishmaniasis (CVL) confirmed by clinical, parasitologic, and serologic examination (data provided by veterinarians).GroupⅣ: a pool of six experimentally infected sandflies (Lutzomya longipalpis)fed with blood from dogs with CVL.GroupⅤ: 18 samples from patients with confirmed tegumentary leishmaniasis (TL) who attended the HCFMUSP complex.Diagnoses were confirmed by clinical manifestations associated with parasitologic visualization of parasites on skin scrapings or skin biopsy samples (data extracted from medical records).Two control groups were included: GroupⅥcomprised 30 samples from healthy blood donors from HCFMUSP provided by the Departamento de Biologia Molecular, Fundação Pró-Sangue/Hemocentro de São Paulo (Certificate of Ethics Presentation,39278514.8.0000.0065).GroupⅦ comprised 47 samples (PB and/or BMA) from patients with signs and symptoms suggestive of VL and/or from areas endemic for leishmaniasis (data extracted from medical records) who had other diagnosed diseases.
2.3.Reference strains
The reference strains of Leishmania spp.used as PCR positive controls were: Leishmania (Leishmania) infantum (MHOM/BR/72/strain46), L.(L.) amazonensis (MHOM/BR/1973/M2269), L.(L.)donovani (MHOM/IN/80/DD8), L.(L.) major (MHOM/1L/80/Friedlin), Leishmania (Viannia) braziliensis (MHOM/BR/75/M2903), L.(V.) guyanensis (MHOM/BR/1975/M4147), L.(V.)shawi (MHOM/BR/2001/M19672), L.(V.) lainsoni (MHOM/BR/81/M6426), and L.(V.) naiffi (MDAS/BR/79/M5533).
2.4.DNA samples from different pathogens
To ensure amplification specificity, DNA samples from different pathogens were tested: Trypanosoma (T.) cruzi (5 samples from patients and Y strain); T.brucei; Mycobacterium (M.) tuberculosis;Toxoplasma gondii (3 different strains); Plasmodium falciparum;Histoplasma capsulatum, and Schistosoma (S.) mansoni.
2.5.Gold standard methods for laboratory diagnosis of human VL
Parasitology and/or serology are the gold standard methods for the laboratory diagnosis of human VL (HVL), recommended by Ministério da Saúde do Brasil (Health Ministry of Brazil).These methods are used to define cases of leishmaniasis.Parasitology is based on microscopy examination of stained smears or cultures prepared from BMA or PB buffy coat samples.Serology is based on ELISA using PB samples or IrK39 using BMA or PB samples.
2.5.1.Parasitology (stained smear technique)
Smears from BMA and/or from PB buffy coat samples were prepared using 5 μL of sample stained with panotic dye (Newprov,Pinhais, Brazil) and analyzed by microscopy (1 000×magnification).
2.5.2.Parasitology (culture technique)
Forty microliters of BMA and/or PB buffy coat were transferred into tubes containing Novy-MacNeal-Nicolle/brain heart infusion medium (DIFCO, Detroit, MI, USA).Aliquots of 10 μL were obtained from the cultures and analyzed by microscopy (400×magnification) once a week, for 30 d.
2.5.3.Serology (IrK39 test)
Whole blood, plasma, or BMA was analyzed using IrK39-IT LEISH (Bio-Rad/DiaMed, Cressier, Switzerland) according to the manufacturer’s pre-established protocols.
2.6.Gold standard method for laboratory diagnosis of CVL
Canine samples were analyzed by parasitologic and serologic examination (ELISA and rapid immunochromatographic dual-path platform tests- Biomanguinhos/FIOCRUZ, Rio de Janeiro, Brazil) to diagnose CVL.These parasitologic and serologic tests are the gold standard for the laboratory diagnosis of CVL.The tests were carried out at the Instituto Adolfo Lutz (Adolfo Lutz Institute), São Paulo,Brazil.
2.7.Gold standard method for laboratory diagnosis of TL
Visualization of parasites on skin scrapings or skin biopsy samples(data extracted from medical records) from 18 patients with TL were carried out at HCFMUSP.
2.8.Molecular techniques performed for groupsⅠ-Ⅶ
Samples from BMA or PB (groupsⅠ,Ⅱ,Ⅲ,Ⅵ, andⅦ), macerated from a pool of sandflies (groupⅣ), and from biopsy samples(groupⅤ) were subjected to PCR.Genomic DNA was extracted from 200 μL of PB or BMA using a QIAamp DNA blood kit(QIAGEN, Hilden, Germany) and a QIAamp DNA tissue kit(QIAGEN), according to the manufacturer’s recommendations.The concentration of the DNA samples was analyzed in a NanoDrop 1 000 spectrophotometer (Thermo Fisher, Boston, MA, USA) and set at 200 ng by PCR.Filter tips with physical barriers were used to minimize the risk of PCR carry over, such as the use of separate work areas (reagent, extraction, and amplification room).All DNA samples from groupsⅠ-Ⅱand Ⅴ-Ⅶwere subjected to PCR of the constitutive human beta-actin gene (B1 and B2) to evaluate that this constitutive protein has not been affected in the tested samples, ensuring the quality of the samples and the inexistence of inhibitors[16].
The procedures for ITS1 PCR (LITSR/L5.8S) and kDNA PCR(RV1/RV2) are described in Table 1[17,18].Products of PCR were visualized on ethidium bromide-stained 2% agarose gels (Agargen,Madrid, Spain) examined on a transilluminator (Alpha Innotech, San Leandro, CA, USA).Twenty microliters of ITS1 PCR products weredigested with 1 U of the restriction enzyme HaeⅢ in 1× buffer(Fermentas, Burlington, ON, Canada), following the manufacturer’s instructions.Restriction fragments were visualized on ethidium bromide-stained MetaPhor agarose gels (Lonza Rockland Inc.,Rockland, Maine, USA) examined on a transilluminator (Alpha Innotech).
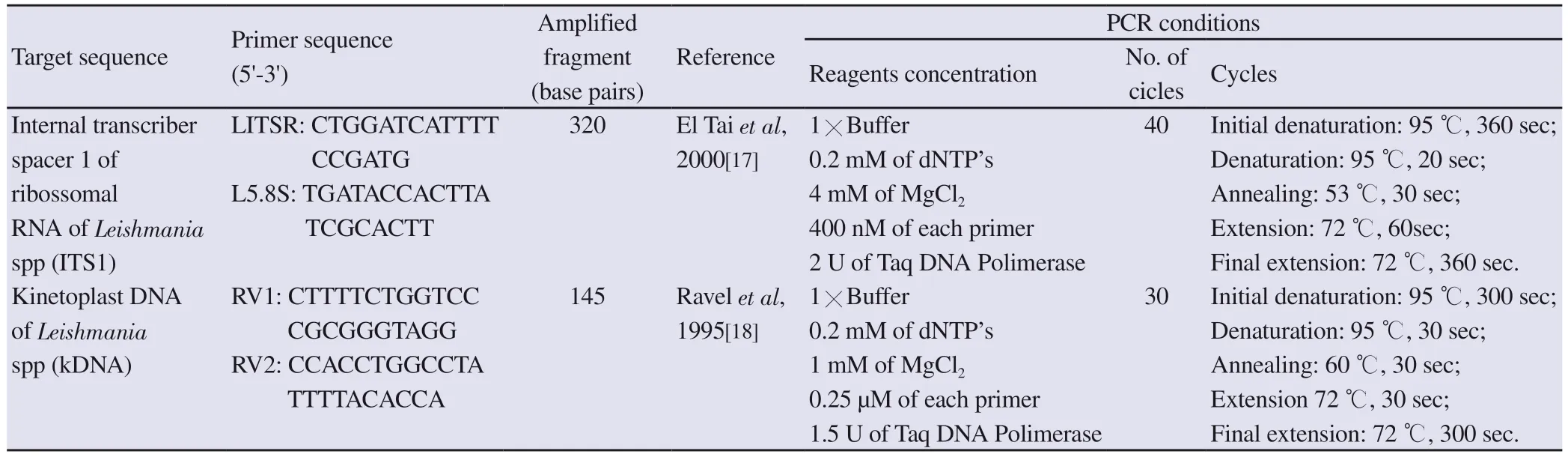
Table 1.Description of target sequences of ITS1-PCR (LITSR/L5.8S) and kDNA-PCR (RV1/RV2), primers, amplified fragments (bp), references and PCR conditions.
2.9.Sequencing
Ten ITS1 PCR amplicons from samples belonging to groupⅠ(VL patients), 2 samples from dogs with confirmed CVL and L.(L.)infantum strain (MHOM/BR/81/M6445), used as a reference, were sequenced.Sequencing reactions were performed on the ABI PRISM 3500 genetic analyzer platform (Thermo Fisher) using the BigDye terminator cycle sequencing kit (Applied Biosystems, Waltham, MA,USA).Then, the electropherograms (sequences), the positive control L.(L.) infantum strain (MHOM/BR/81/M6445), and the reference sequence from the region of interest (KF985171.1) retrieved from GenBank (https://www.ncbi.nlm.nih.gov/genbank) were manually edited using the BioEDIT sequence alignment editor.The alignment of sequences was examined using the codon code aligner and compared using the basic local alignment search tool (BLAST)sequence analysis tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and the Sequencher 4.1.4 program (Genes Code Corporation, Ann Arbor,MI, USA).
Data analysis was performed to determine the sensitivity,specificity, positive and negative predictive values, and efficiency of ITS1 PCR and kDNA PCR.The kappa index was used to evaluate the proportion of agreement, in addition to that expected by randomness, between the two molecular tests and the gold standard examinations, adopted as reference in the present study(parasitologic tests on BMA and PB samples, IrK39, and clinical and epidemiologic data).The confidence interval was 95% based on the estimated standard kappa error of samples and their Z score.A P value <0.05 was considered significant.The STATA program,version 13.0 (Stata Corp LP, College Station, TX, USA) was used for the statistical analyses.
3.Results
3.1.Gold standard methods for laboratory diagnosis of HVL,CVL, and TL
3.2.Molecular techniques for groupsⅠ-Ⅶ
All samples from groupsⅠ-Ⅱ,Ⅴ-Ⅶ successfully amplified the 520 base pair (bp) fragment from the beta-actin gene, which ensured the absence of amplification inhibitors; this constitutive protein was unaffected in all samples tested.
ITS1 (LITSR/L5.8S) and kDNA (RV1/RV2) generated fragments of 320 bp (Figure 1) and 145 bp (Figure 2), respectively.They were positive in 97.6% (80 of 82) and 92.7% (76 of 82) of the samples from groupⅠ(82 patients with HVL), respectively (Table 2).

Table 2.Results of the gold standard exams and molecular techniques of the 7 groups.

Figure 1.Agarose gel electrophoresis (2%) of ITS1-PCR (320 base pairs) from positive samples of patients with visceral leishmaniasis(VL).(100) 100 bp DNA ladder.(R) Negative control of reagents room.(E) Negative control of extraction room.Lanes 1, 2 and 3-samples of patients with VL.(PC) L.(L.) infantum positive control.

Figure 2.Agarose gel electrophoresis (2%) of kDNA(RV1-RV2)-PCR(145 bp) from positive samples of patients with visceral leishmaniasis (VL).(50) 50 bp DNA ladder.(R) Negative control of reagents room.(E) Negative control of extraction room.Lanes 1, 2, 3 and 4 samples of patients with VL.Lanes 5 and 7-samples of patients with VL but negative in kDNA (RV1-RV2).(PC) L.(L.) infantum positive control.
After ITS1 PCR RFLP (HaeⅢ) analysis of the 80 positive samples,52.5% (42 of 80) generated three fragments of 180, 70, and 50 bp,corresponding to the pattern of L.(L.) infantum (Figure 3).

Figure 3.Metaphor agarose gel electrophoresis (4%) of ITS1-PCR-RFLP for the evaluation of profiles obtained from samples of patients with visceral leishmaniasis (VL).(100) 100 bp DNA ladder.(LA) L.(L.) amazonensis(190 and 140 bp), (PC) L.(L.) infantum (180, 70 and 50 bp).Lanes 1 and 2-samples from patients with VL.(LM) L.(L.) major (200, 140 bp).(LB) L.(V.) braziliensis (160, 150 bp).
Of the 16 samples analyzed from groupⅡ(VL patients under treatment), 37.5% (6 of 16) presented positive results for at least one of the molecular tests.ITS1 PCR detected 12.5% (2 of 16)of the samples from groupⅡ, and these 2 samples generated no profiles by ITS1 PCR RFLP, because the 320 bp fragment remained unrestricted.Of the 16 samples, 25.0% (4 of 16) were positive by kDNA PCR (RV1/RV2).
ITS1 PCR and kDNA PCR diagnosed 85.7% (12 of 14) from group Ⅲ(CVL).According to the results obtained by kDNA, these 12 samples belonged to Leishmania subgenus.ITS1 PCR RFLP showed that 83.3% (10 of 12) of the canine samples contained parasites with profiles similar to L.infantum.
Pool samples of DNA from six infected sandflies (groupⅣ) were tested by ITS1 PCR and kDNA PCR, and both systems yielded amplifications (Table 2).ITS1 PCR products were analyzed by RFLP, generating a profile similar to that of L.infantum.
In groupⅤ(patients with TL), 72.2% (13 of 18) of the samples analyzed by ITS1 PCR were positive (Table 2) and 69.2% (9 of 13)showed profiles corresponding to a Viannia complex by ITS1 PCR RFLP.
Regarding the specificity of the ITS1 PCR, there was no amplification with DNA samples from other pathogens, whereas kDNA PCR amplified DNA from S.mansoni.When kDNA PCR was carried out with DNA from reference strains, L.(L.) amazonensis and L.(L.) infantum were amplified.These two species belong to Leishmania subgenus.
学生在明确自己的学习任务后,带着问题进入教师设置的情境中去,查阅相关资料,寻找解决问题的方法。通过小组讨论区的在线论坛,小组成员在讨论板上通过发帖的方式与小组成员讨论或者向老师请教,教师进行实时辅导、答疑、教学交流。在解决了交互中的差异后,正副组长总结结论,记录备案,或制作PPT展示,以达到团队合作学习的目的。
Thirty samples from groupⅥ (blood donors) and 47 from groupⅦ(other diseases) were negative by ITS1 and kDNA molecular tests(Table 2).As shown in Table 3, when PCR results were compared with the gold standard for HVL, near perfect agreement was observed, with a kappa index >0.80 and a P value <0.001.
For samples from patients with confirmed HVL, 90.2% (74 of 82) showed agreement between ITS1 and kDNA PCR.There was disagreement in 9.8% (8 of 82) of the samples; two were negative by ITS1 PCR and positive by kDNA PCR, and six were positive by ITS1 PCR and negative by kDNA PCR.Five of these six samples presented a profile corresponding to L.(L.) infantum by ITS1 PCR RFLP.
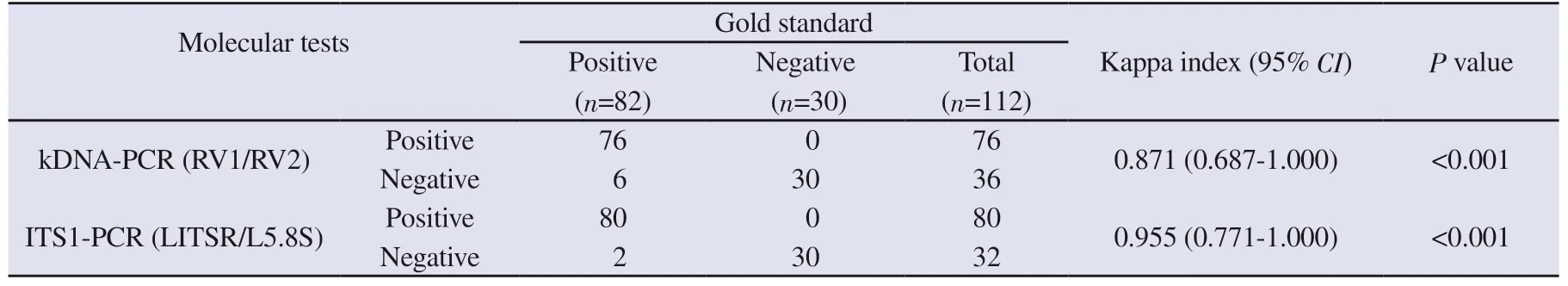
Table 3.Comparative analysis of molecular diagnosis by ITS1-PCR and kDNA-PCR using samples from confirmed cases of visceral leishmaniasis (GroupⅠ)and samples from healthy individuals (GroupⅥ).
3.3.Sequencing
It was possible to analyze the results of nine of 12 samples sequenced (eight from groupⅠand one canine sample with CVL from groupⅣ) and 100% (9 of 9) showed 99% similarity with the reference sequence (KF985171.1).
3.4.The proposed algorithm
Based on our findings (parasitologic, immunologic, and molecular tests), an algorithm is proposed for processing samples until the species is defined (Figure 4).According to our proposal, the parasitologic and immunochromatographic diagnoses should be considered before the molecular tests, and kDNA PCR, for the determination of Leishmania subgenus, should be done after a negative ITS1 PCR.A positive ITS1 PCR must be followed by ITS1 PCR RFLP to define the agent causing VL.
4.Discussion

Figure 4.Algorithm proposed for processing samples at the species level from patients with suspected VL.VL=visceral leishmaniasis; Irk39=immunochromatographic test rk39; BM=bone marrow; PB=peripheral blood; PT=parasitological tests; NEG=Negative; POS=Positive; bp=base pairs.
A case of VL is confirmed when, even before clinical suspicion;a positive laboratory diagnosis is demonstrated by parasitologic or serologic tests (indirect immunofluorescence ELISA, or immunochromatographic tests using recombinant antigens)[14].In our study, 76.3% (61 of 80) of patients with VL (groupⅠ) were positive by parasitologic examination of smears from BMA/PB stained with Panotic dye.Koltas et al.[3] reported that 20.8% (10 of 48) and 80.0%(8 of 10) of smears and cultures of BMA from children with clinical suspicion of VL were positive, respectively.Sensitivities of 98.0%,87.9%, and 72.7%, respectively, were obtained by kDNA PCR,smears, and cultures of samples of patients with TL in a study by Rasti et al[4].Although parasitologic tests demonstrate the presence of the parasite in the samples, they can not define the causative agent of VL, because it is not possible to distinguish the parasite by microscopic analysis in both techniques[3,19].
Several studies report high sensitivity and specificity of IrK39[20].Although the test is considered specific for the diagnosis of VL[21],there are reports of false-positive results[22,23].IrK39 was positive in 68.8% (55 of 80) of the samples from groupⅠ, which comprises patients with confirmed VL.The samples that were positive for IrK39 included some patients coinfected with Leishmania and HIV,and 60.0% (15 of 25) of these samples were positive in IrK39.This result corroborates findings from other authors in which IrK39 had decreased sensitivity due to the presence of immunodeficiency and achieved 45% sensitivity[21].
Although VL is usually caused by L.(L.) infantum in Brazil,it is important to identify the species responsible for the VL to allow suitable treatment.It is also necessary to identify species in epidemiologic surveys and to define the species responsible for atypical symptoms in patients with Leishmania/HIV coinfection or even in immunocompetent patients[3,24].Using molecular approaches, such as real-time PCR or conventional PCR, it is possible to identify the species involved in the leishmaniasis infection.The sensitivity of real-time PCR is superior to conventional PCR, in addition to presenting other advantages[24,25]; however it is an expensive technology.Because leishmaniasis is a neglected disease, this technology is not available to the public health system in Brazil, which explains our choice of conventional PCR.With conventional PCR, depending on the target chosen in DNA, and/or performing RFLP, it is possible to define the species of the agent responsible for VL[25].Therefore, ITS1 sequences were chosen because they are composed of highly conserved regions, allowing their use in PCR for diagnostic purposes, and they have polymorphic regions that can be used in RFLP assays to determine the species.Also, having kDNA as a target and using the RV1/RV2 primer pair,it is possible to demonstrate the Leishmania subgenus present in the infection.When analyzing the sensitivity of the PCR with the primer pair (LITSR and L5.8S) target in the ITS1 region of the DNA, the sensitivity was superior (97.6%) to that of kDNA (92.7%) when tested in samples from patients with VL (groupⅠ); however, this difference was not significant.These sensitivities (ITS1 and kDNA)were higher than the sensitivity for parasitology (76.3%) and IrK39(68.8%) for samples from patients with VL.On the other hand, in a study with 431 blood donors from the state of Ceará (Brazil), ELISA detected more positive samples (13.2%, 57 of 431) than kDNA PCR(K20/K22) (4.6%, 20 of 431)[26].Khan et al.[14] reported sensitivities of 98.4% (60 of 61) and 96.7% (59 of 61) with IrK39 and ITS1,respectively.
In contrast to our findings with ITS1, Beldi et al.[27] found low sensitivity (63.9%, 23 of 36) in samples from patients with VL in Algeria; however, the authors used smears of BMA to obtain the DNA of the parasite.Koltas et al.[3] reported a sensitivity of 90% (9 of 10) with ITS1 in samples from patients with VL.
Regarding specificity, ITS1 did not amplify any DNA in pathogen samples (T.cruzi, T.brucei, M.tuberculosis, Toxoplasma gondii,Plasmodium falciparum, Histoplasma capsulatum, and S.mansoni)in our study.Ozerdem et al.[28] also reported 100% specificity with ITS1.Some studies have tested ITS1 with strains of M.tuberculosis,M.leprae, S.mansoni, Wuchereria bancrofti, and T.cruzi, and also without non-specific amplification, demonstrating the importance of this target in terms of analytical specificity[28,29].In our findings,kDNA (RV1/RV2) was considered acceptable to define the Leishmania subgenus, because there was amplification of DNA from a reference strain of L.(L.) amazonensis, which belongs to the same subgenus of L.(L.) infantum.Solcà et al.[30] demonstrated the amplification of kDNA (RV1/RV2) for L.(L) major and L.(L.)amazonensis.Therefore, amplification of the subgenus Leishmania using RV1/RV2 primers was demonstrated in these studies.
Multilocus enzyme electrophoresis is the gold standard technique for the identification of Leishmania species.However, it is an expensive and laborious method that requires culturing of the parasite before its execution[8].We have proposed an algorithm for the molecular diagnosis of VL-specific species using the primer pair LITSR/L5.8S (ITS1 PCR) to identify the species.Thus, we used RFLP as an alternative to multilocus enzyme electrophoresis,with restriction enzyme HaeⅢ, on the products of ITS1 PCR.The ITS1 PCR RFLP technique is widely used in studies involving leishmaniasis in both the Old and New World[19,25,31].It is known that the causative agent found in the American continent is L.(L.) infantum, which belongs to a donovani complex[2].Of the 80 positive samples that amplified in the ITS1-PCR, 52.5% (42 of 80)demonstrated a profile similar to that of L.(L.) infantum by ITS1 PCR RFLP.However, ITS1 PCR diagnosed 85.7% of the canine samples (12 of 14) from groupⅢ, and ITS1 PCR RFLP showed that 83.3% (10 of 12) of dog samples contained parasites with profiles similar to L.(L.) infantum.The distinction between species of the Viannia and Leishmania subgenus was very clear, even without the use of high-resolution gel.But the different sensitivities obtained using ITS1 PCR RFLP between humans (52.5%, 42 of 80 in group Ⅰ) and dogs (83.3%, 10 of 12 in groupⅢ) can be related to the low parasitic load in humans[31], explaining the lack of species definition using ITS1 PCR RFLP in 48.8% (39 of 80 ) of our positive human VL samples (groupⅠ).
In the study by Hijjawi et al.[32], ITS1 PCR RFLP was able to identify the species responsible for TL in Jordan in 28 of the 30 positive samples in ITS1 PCR.Monroy-Ostria et al[19], when testing skin lesions in patients from Mexico with ITS1 PCR RFLP (HaeⅢ),obtained different restriction profiles for the species L.(L.) mexicana,L.(L.) amazonensis, and a third profile that grouped the species L.(V.) panamensis, L.(V.) guianensis, and L.(V.) braziliensis.
Of the 18 samples belonging to groupⅤ(patients with TL), 72.2%(13 of 18) were positive in ITS1.The 13 positive samples showed an electrophoretic profile similar to that found for L.(V.) braziliensis,L.(V.) lainsoni, L.(V.) shawi, and L.(V.) guyanensis.Amro et al.[33]have described possible inhibition or failure of ITS1 amplification in samples from TL patients in the Old World.
These two systems (ITS1 and kDNA) complemented each other in our study; they diagnosed 100% of the samples belonging to the Leishmania genus.In addition, kDNA defined Leishmania subgenus in 92.7% of the samples and more specifically, L.(L.)infantum was identified by ITS1 PCR RFLP in 52.5% (42 of 80) of positive samples.Based on these findings, we suggest an algorithm for the molecular diagnosis of VL, which must first consider the parasitologic and immunochromatographic diagnosis.This molecular diagnosis can combine two PCR target systems: ITS1 and kDNA.ITS1 complemented with RFLP can define the L.(L.) infantum species, corroborating the IrK39 findings (donovani complex).Also,for samples with negative results of ITS1, kDNA target (RV1/RV2)should be used to determine, at least, the Leishmania subgenus.
 Asian Pacific Journal of Tropical Medicine2020年2期
Asian Pacific Journal of Tropical Medicine2020年2期
- Asian Pacific Journal of Tropical Medicine的其它文章
- Dengue virus infections and anti-dengue virus activities of Andrographis paniculata
- Human leukocyte antigen class-Ⅱ DRB1 alleles and Giardia lamblia infection in children: A case-control study
- In vitro biological activities of aqueous extracts of Tetrapleura tetraptera (Schumach.& Thonn.) taub.and Aframomum citratum (C.Pereira) K.Schum from three Agroecologic Zones in Cameroon
- Predicting the number of visceral leishmaniasis cases in Kashgar, Xinjiang, China using the ARIMA-EGARCH model
- Co-existence of renal hydatid cyst and renal cell carcinoma in one kidney: A case report
- First evidence of Bartonella phoceensis and Candidatus Mycoplasma haemomuris subsp.ratti in synanthropic rodents in Malaysia
