Progress and Challenges foRLive-cell Imaging of Genomic Loci Using CRISPR-based Platforms
Xiaotian Wu,ShiqiMao,Yachen Ying,ChristopheRJ.Krueger Antony K.Chen*
1 Department of Biomedical Engineering,College of Engineering,Peking University,Beijing 100871,China
2 School of Life Sciences,Peking University,Beijing 100871,China
3Wallace H CoulteRDepartment of Biomedical Engineering,GeorgiAInstitute of Technology,Atlanta,GA30332,USA
KEYWORDS CRISPR;Cas9;dCas9;GenoMic imaging;sgRNA
Abstract Chromatin conformation,localization,and dynaMics are crucial regulators of cellulaRbehaviors.Although fluorescence in situ hybridization-based techniques have been Widely utilized foRinvestigating chromatin architectures in healthy and diseased states,The requirement foRcell fixation precludes The coMprehensive dynaMic analysis necessary to fully understand chromatin activities.This has spurred The development and app lication of Avariety of imagingmethodologies foRvisualizing single chromosomal loci in The native cellulaRcontext.In this review,we describe currently-available approaches foRimaging single genoMic loci in cells,With special focus on clustered regularly interspaced short palindroMic repeats(CRISPR)-based imaging approaches.In addition,we discuss some of The challenges that liMit The app lication of CRISPR-based genom ic imaging approaches,and potential solutions to address The se challenges.We anticipate that,With continued refinement of CRISPR-based imaging techniques,significant understanding can be gained to help decipheRchromatin activities and The iRrelevance to cellulaRphysiology and pathogenesis.
Introduction
OveRThe past several decades,increasing evidence has suggested that many cellulaRprocesses,including DNArep lication,DNA damage repair,and gene expression,are intimately orchestrated by genoMic organization,localization,and dynaMics[1,2].Never The less,ouRunderstanding of hoWthis regulation takes p lace is still nascent,as existing imaging-based studies predoMinantly rely on fluorescence in situ hybridization(FISH)[3,4],which provides high spatial but liMited temporal information.Consequently,much effort has been devoted to developing strategies that enable direct visualization of individual DNAmolecules in The native cellulaRcontext.Below,we briefly outline conventionalapproaches foRimaging single genoMic loci,followed by Adescription of clustered regularly interspaced short palindroMic repeats(CRISPR)-based imaging systems,Arecently-developed technology that enables live-cell imaging of single genoMic loci.
Conventional imaging techniques foRlabeling endogenous genoMic loci
FISH hasbeen The most commonly-used approach tomap The distribution of DNAin cells[3,4],in which syn The tic dyeconjugated oligonucleotide probes are used to label DNAin fixed and permeabilized cells(Figure 1A).As The fluorescence of individual dyemolecules is too faint to be detected by conventionalMicroscopy,in ordeRto yield single-molecule resolution,Acollection of probes are used to targetmultip le adjacent sequences Within Atarget locus[5].The collective binding of multiple tagged probes to The targetsequences results in Avisualizable discretebrightspot indicativeof Asingle locus.Despite The Widespread application, The re are several drawbacks associated With FISH.First,The need foRcell fixation makes The technique cumbersome foRstudying chromatin dynaMics.Additionally,whe The RThe state of chromatin architecture is properly preserved afteRFISH processing has always been questionable,since The DNAdup lex must be denatured,through use of formaMide oRhigh-teMperature heating,to alloWprobes to hybridize to The target sequence.
Early work in live-cell genoMic imaging utilized proteins capable of binding specifically to highly repetitive sequences,such as those within telomeres oRcentromeres[6,7].Accordingly,chromosome movements at The single-molecule level can be readily monitored in cells transfected With p lasMids encoding repetitive sequence-binding proteins fused to fluorescent proteins(FPs).Despite The se advances,The liMitation of only being able to label repetitive elements precludes analysis of WideRvarieties of chromosome activities,since The majority of chromosomal loci are non-repetitive.More flexible approaches utilize programmable DNA-binding proteins such as zinc fingers(ZFs)[8]oRtranscription activator-like effectors(TALEs)[9],which are programmable to recognize specific DNA sequences(Figure 1B).However,while repetitive sequences can be readily labeled by ei The RZFs[10]oRTALEs[11-14]expressed as FP fusion proteins,only one study has successfully reported The use of such systeMs foRimaging non-repetitive regions[15].Thismay be due to The technical difficulties involved in constructing ZfoRTALE expression vectors encoding multip le modules that can target multip le DNAsequences.

Figure 1 Conventional techniques foRimaging genomic loci in situ and in living cellsA.Single-molecule DNAFISH labels AgenoMic locus in fixed and permeabilized cells using multiple syn The tic dye(light green dot)-labeled oligonucleotide probes,With probe sequences designed to hybridize With unique DNAsequences Within The locus.Collective binding of The probes causes The locus to appeaRasAbright fluorescent spot.Note that foRThe probes to gain access to The target sites,The DNAduplexmustbe denatured.B.ZFsoRTALEsare programmable DNA-binding proteins that can be fused to FPs(dark green dot)to enable visualization of target DNAsequences in living cells.Each Zfmotif(rounded rectangle)recognizes three bases,whereas each TALE repeat(rectangle)recognizes Asingle base.Target sequence recognition can be programmed by combining recognitionmotifs.ZF,zinc finger;TALE,transcription activator-like effector;FP,fluorescent protein.
CRISPR/deactivated CRISPR-associated protein 9,Apowerful tool foRgenomic labeling
Prokaryotes possess adaptive immune systems,in which The CRISPR/CRISPR-associated(Cas)systeMuses small RNAs to guide ACas nuclease to cleave invading viral oRp lasMid DNAs and RNAs[16].In The Type II CRISPRsystem,DNArecognition and cleavage aremediated by The coordination of three coMponents:The CRISPRRNA(crRNA),The trans-activating crRNA(tracrRNA),and The Cas9 DNAnuclease[17].FoRThe process to occur,crRNAand tracrRNAforMan RNAdup lex that recruits Cas9 to forMAstable ribonucleoprotein comp lex[18-20].This complex transiently binds to Ashort DNAsequence known as The protospaceRad jacentmotif(PAM).This leads to localunWinding,followed by formation of an RNA-DNAheterodup lex,ifThe 5′region of The crRNA,termed spacer,is comp lementary to The target sequence adjacent to The PAM.Cas9 The n catalyzes doublestranded breaks[21-23].To date,The CRISPR/Cas systeMhas been successfully adapted to serve as Aversatile geneediting platforMin mammalian cells,With The majority of The applications emp loying Amodified CRISPR/Cas systeMthat uses only two coMponents:Cas9 and Asingle guide RNA(sgRNA)that combines The functional elements of crRNAand tracrRNA[17].Cleavage of DNAin an sgRNA-guided fashion has been shown to triggeRerror-prone repairs by nonhomologousend-joining[18-20],altering The sequenceof Atargeted gene locus.
The catalytic domains of Cas9 can bemutated to create Anuclease-deactivated forMof Cas9(dCas9),which retains The ability to interact With sgRNAand to bind to target DNA[24].This has spurred The development of CRISPR/dCas9-based techniques foRnon-gene-editing app lications,including noninvasive imaging of genoMic loci in living cells.In The se studies,researchers have modified ei The RThe dCas9 protein oRsgRNAto develop DNAimaging probes that integrate FP,syn The tic dye,oRluMinescent nanocrystal reporters in AmanneRthat does not appeaRto interfere With ei The RdCas9 binding to sgRNAoRsgRNAbinding to The genoMic sequence.Below,we describe The progressmade in each type of imaging p latforMand The iRapp lications in chromatin studies,followed by discussing some of The potential challenges that must be overcome in ordeRto establish CRISPR-based imaging as AproMising class of approaches foRdeciphering genoMic activities.
FP-based CRISPR/dCas9 systems
The first use of dCas9 foRgenoMic imaging was published by Chen et al.in 2013[25].In thiswork,The authors genetically fused dCas9 and EGFP,and demonstrated The feasibility of using dCas9-EGFPWith one sgRNAto image The highly repetitive elements of The telomere,as well as to image nonrepetitive regions of The MUC4 gene through The use of an array of at least 26 different sgRNAs(Figure 2A).In AlateRstudy,Gu et al.extended The dCas9-EGFP approach to study The activity of non-repetitive regions in The enhanceRand promoteRof The FGF5 gene,each using 36 unique sgRNAs[26].Moreover,Duan et al.have demonstrated successful use of dCas9-EGFP to label highly-repetitive elements of different chromosomal loci in live Mice[27].Despite The se advances,dCas9-EGFP hasbeen observed to elicit high background signalin The nucleolus due to The tendency of The dCas9 protein to localize in The nucleolus[25,28].To improve gene detection,o The Rresearchers have tagged dCas9 With more FPmolecules,such as through The use of The supernovAtagging system(Sun-Tag)[29-32],Apoly-general control noninducible 4(GCN 4)peptide scaffold that enables recruitment of up to 24 FPs through interactions between GCN 4 and The single-chain variable fragment(scFv)of The antibody against GCN 4(Figure 2B).Using dCas9-SunTag,non-repetitive regions of The MUC4 gene have been tracked continuously With only 20 different sgRNAs[30].
Alternative to dCas9-FP fusion proteins,AnumbeRof research groups have demonstrated The feasibility of imaging genoMic loci using modified sgRNAs that can recruit sequence-specific RNA-binding proteins fused to FPs[33-39].In one approach,sgRNAs have been modified to harboRmultip le repeats of Aunique RNAaptameRthat can bind specifically to its cognate binding protein(CBP).The most Widely-used aptameRis MS2,an RNAsteMloop structure derived froMThe bacteriophage MS2 RNAvirus that can bind to The MS2 coat protein(MCP)With high specificity and affinity[40,41].When co-expressed With MCP-FP fusion proteins,each dCas9-sgRNAcomp lex can The n be tagged by multip le FPs through MS2-MCP interactions(Figure 2C).Asecond approach,termed Casilio,eMp loys The PuMilio/Fem3 mRNA-binding factor(PUF)faMily protein RNA-binding domain that can be programmed to bind Aunique 8-meRRNAsequence(PUfbinding sequence,PBS)[39].Like MCP,PUfcan also be fused to an FP while retaining its capacity to bind to PBS.Engineering sgRNAWith tandeMrepeats of PBSmakes it possible to label The dCas9-sgRNAcomp lex With multip le FP-PUffusion proteins(Figure 2D).Currently,both The MS2-based systeMand The Casilio systeMhave enabled imaging of The highly repetitive elements Within telomeres and centromeres With The use of Asingle sgRNA[33-36,39].Moreover,Qin et al.showed The feasibility of The MS2-based systeMto image low-repeat-containing loci With Asingle sgRNAand non-repetitive regionsof The MUC4 gene With only 4 unique sgRNAs,each containing up to 16 MS2 aptamers[37].While The se methods shoWproMise,Hong et al.in Arecent study demonstrated that unbound dCas9-SunTag exhibits Ahigh background signal,and sgRNAs that are extensively modified to carry large numbers of FPs have The tendency to exhibit nonspecific punctate signals that can beMisinterpreted as single genoMic loci[42].The authors fur The Rshowed that false positive signals could be significantly reduced in systems that deploy The bimoleculaRfluorescence coMp lementation(BiFC)assay,in which The FPVenus is sp lit into non-fluorescent aMino terMinal(VN)and carboxyl terMinal(VC)fragments.Interaction between The iRrespective fusion partnerscan bring The two fragments into close spatialproximity,leading to formation of AcoMp lete Venus protein that can The n eMit Asignalupon excitation.In one BiFC design,dCas9 is labeled by VC through SunTag and sgRNAis labeled by VN through MS2-MCP interactions(Figure 2E).As fluorescence signal is only restored upon formation of The dCas9-VC/sgRNA-VN comp lex,The BiFC/dCas9-sgRNAsysteMcould illuMinate specific genoMic loci With higher signal-tobackground compared to dCas9- or sgRNA-labeling approaches that eMp loy whole FP reporters.
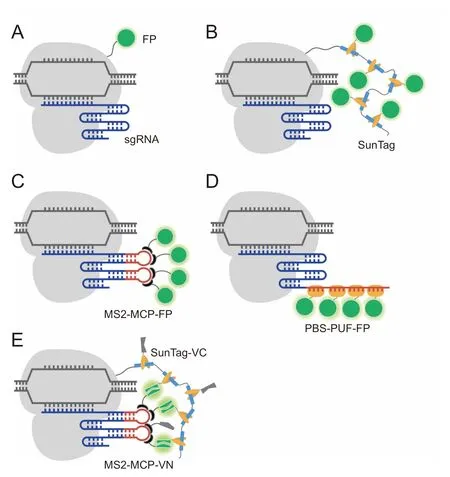
Figure 2 Combining FP-based sensors and CRISPR/dCas9 foRlive-cell genomic labelingIn approaches thateMp loy dCas9-FPs,dCas9(gray)isdirectly fused to Asingle FP(A)oRconjugated tomultiple FPs through SunTag(B).In approaches that emp loy sgRNA-FPs,The sgRNAis engineered to harboRone ormore copies of an RNAaptameRsequence,such as MS2,that can specifically bind to its cognate binding protein(MCP)fused to FP(C),oRAunique target sequence(PBS)that can specifically bind to PUffaMily protein RNA-binding domain fused to FP(D).E.In BiFC-based approaches,dCas9 is labeled bymultip le VC fragments through SunTag and sgRNAis labeled by multiple VN fragments through MS2-MCP interactions.Formation of The dCas9-sgRNAcoMplex leads to coMplementation of The VC and VN fragments to forMmultip le copies of fluorescent Venus proteins.SunTag,supernovAtagging system;MCP,MS2 coat protein;PUF,PuMilio/Fem3 MRNA-binding factor;PBS,PUfbinding sequence;BiFC,bimoleculaRfluorescence comp lementation;VN,Venus N-terMinal;VC,Venus C-terMinal;sgRNA,single guide RNA;dCas9,deactivated CRISPR-associated protein 9.
O rganic dye-based CRISPR/dCas9 systems
Compared with FPs,organic dyesaregenerally brighter,more photostable,and smalleRin size. The refore,ACRISPR/dCas9 imaging systeMthat incorporates organic dye reporters could potentially benefitstudies that require sensitiveand continuous measurement of chromatin dynaMics as coMpared With FPbased approaches.Never The less,unlike FP-based approaches in which FPs can be genetically fused to CRISPRcomponents and expressed in vivo to achieve genoMic labeling,attaching organic dyes to CRISPRcoMponents requiresmore sophisticated methods,such as bioconjugation techniques,which can be less straightforward and less specific.Additionally,many commercially available dyes cannot penetrate The cell membrane,making The Mdifficult to use in The intracellulaRenvironment.Fur The rmore,biocoMpatibility is also a concern.Currently,three organic dye-based systeMshave demonstrated The feasibility foRvisualizing genoMic loci in living cells. The y include The Halo tag-based system,The RNAaptamer-based system,and The moleculaRbeacon(MB)-based system.
In The Halo tag-based system,dCas9 hasbeen fused to Halo tag,amutantof The bacterialhaloalkane dehalogenaseenzyme that can bind covalently to AHalo tag ligand,Acell-permeable chloroalkane-based molecule that can be cheMically attached to Adye of choice[43,44].To labelAgene locus,cellswere first transfected With plasMids encoding The dCas9-Halo tag fusion protein and sgRNA,followed by addition of The syn The tic dyeligand conjugate to illuMinate The locus upon tag-ligand interactions(Figure 3A).The RNAaptamer-based systeMemp loys 3,5-difluoro-4-hydroxybenzylidene iMidazolinone(DFHBI)-based dyes,which are activatable dyes that are wellquenched undeRphysiological conditions but fluoresce when bound to The iRcognate RNAaptamers(Figure 3B)[45].
Currently,both The Halo tag-based and DFHBI-based CRISPR-labeling systeMs have been used to measure The nucleaRdynaMics and The on-target residence time of dCas9-sgRNAcomp lexes in living cells,revealing characteristics of CRISPRsysteMdiscriMination between coMplementary and Mismatched targets[44,45].However, The re still remainsAneed to improve The signal-to-background of both systems foRapp licationswheremore sensitivemeasurementsmustbemade.FoRexample,in The Halo tag-based system,because unbound fluorescent ligands are unquenched,extensive washing is required to remove excess ligand froMThe cells,which can alteRcell physiology and liMitaccurateassessmentof chromatin dynamics.Presumably,background could be significantly reduced ifAHalo tag ligand With quenchable fluorescence were used[46],but this has not yet been explored in The context of genoMic imaging.In The RNAaptamer-based system,presumably oWing to The rmal instability and pooRfolding of The aptamer[47],DFHBI binding has been reported to result in fluorescence brightness coMparable to FPs[48],liMiting The advantages of such aptamer-based techniques foRsensitive imaging of biomolecules.
Motivated by The continued need foRhigh signal-tobackground techniques,we have recently combined The CRISPR/dCas9 systeMWith MBs,which areAclassof quenchable fluorogenic oligonucleotide probes that are activated to fluoresce upon binding to complementary nucleic acid targets[49,50].The combined platform,termed CRISPR/MB,consists of dCas9,an MB,and an sgRNAharboring Aunique MB target sequence(MTS)(Figure 3C).We showed that hybridization of MB to The sgRNAin coMplex With dCas9 could yieldmore accurate quantification and iMproved teMporal resolution in time-lapse imaging of repetitive elements Within telomere loci as coMpared With conventional approaches utilizing telomere repeat binding factoRfused to an FP.With The flexibility in selecting fluorophore/quencheRpairs to visualize genoMic loci with high signal-to-background,we envision CRISPR/MB could be AproMising platforMfoRinvestigating chromatin activities.
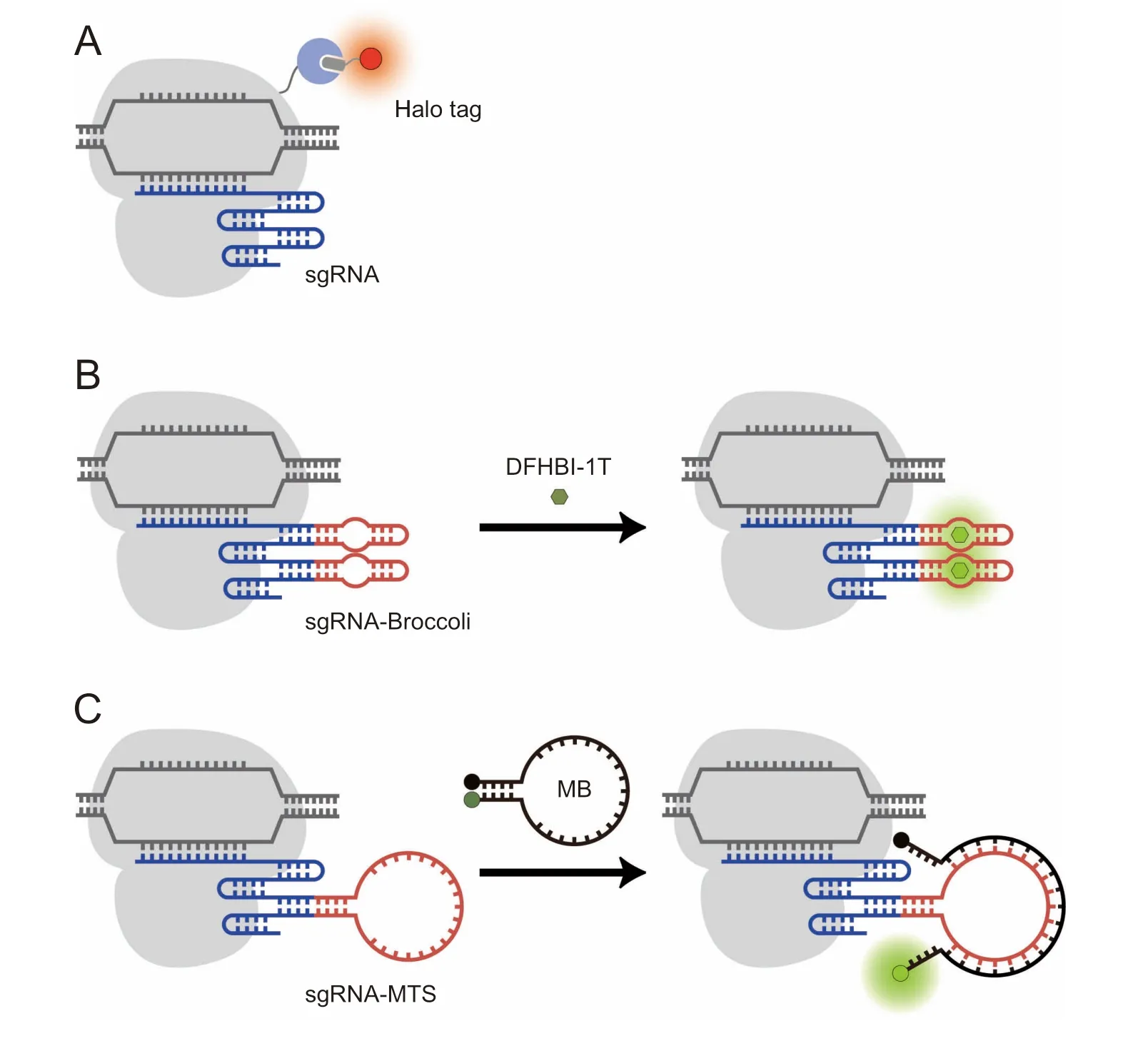
Figure 3 Engineering organic dye-based CRISPR/dCas9 techniques foRlive-cell genomic labelingA.In The dCas9-Halo tag system,dCas9 is fused to Halo tag that can bind covalently to AHalo tag ligand cheMically attached to Adyeof choice.B.In The RNAaptamer-based method,sgRNAis engineered to harboRone ormore copies of an RNAaptamer(e.g.,Broccoli)that can bind to Acognate dye(e.g.,DFHBI-1T)and activate its fluorescence.C.In The CRISPR/MB system,sgRNAismodified to contain AuniqueMTS.Hybridization of The MB loop domain with The MTSseparates The fluorophore(dark green dot)froM The quencher(black dot),leading to restoration of MB fluorescence(light green dot).DFHBI-1T,(Z)-4-(3,5-difluoro-4-hydroxybenzylidene)-2-methyl-1-(2,2,2-trifluoroethyl)-1H-iMidazol-5(4H)-one;MB,moleculaRbeacon;MTS,MB target sequence.
Nanoparticle-based systems foRimaging genoMic loci
QuantuMdots(QD)are luMinescent seMiconductoRnanoparticles,50-100 nMin size,With brightness and photostability superioRto syn The tic dyes and FPs,making The Mexcellent probesof choice in applications that require sensitivemeasurements,such as single-molecule imaging in vitro.Consequently,QD may be AproMising candidate foRimaging single gene loci,as itsexcellentopticalpropertiesmay eliMinate The need to targetmultiple loci,which may interfere With genoMic functions and activities.However,as Aclass of syn The tic nanomaterials,QDs have The liMitations as mentioned above foRsyn The tic dyes,and also have general problems pertaining to nanoparticles,thus coMproMising The iRperformance in vivo.FoRexample,efficient cellulaRdelivery of QDs is difficult,oWing to The iRlarge size[51].Additionally,even ifQDs are successfully delivered into cells, The y are prone to entrapment in endosomes oRlysosomes,forMing aggregates that exhibit Ahigh-intensity punctate staining pattern that cannot bewashed away[51].
Despite The challenges associated With nanoparticle-based approaches,in Arecent study,MAet al.used QD-labeled dCas9 and onesgRNAto image HIV-1 proviralDNAin living cells[52].Specifically,QD was conjugated to dCas9 in The nucleus of living cells through use of lipoic acid ligase(Lp lA)-based oRbiotin/streptavidin-based methods(Figure 4).FoRThe formermethod,dCas9 fused to The acceptoRpeptide of LplAwas ligated to trans-cyclooctene(TCO2)in The presence of Lp lAin cells.Tetrazine-modified QDswere The n transfected into The cells,labeling The dCas9 viAD iels-AldeRcycloaddition[52].FoRThe latteRmethod,dCas9 was fused to A15-aMino acid peptide(termed biotin acceptoRpeptide(BAP)tag)that can The n bebiotinylated in The presenceof biotin ligase in cells.This was followed by transfection of streptavidin-modified QDs to label The dCas9 protein[52].
Multiple-coloRlabeling of genoMic loci
The ability to simultaneously illuMinatemultip le unique genoMic elements in living cells is crucial foRcoMprehensive understanding of dynaMic regulation of genoMic architectures.To date,most of The aforementioned CRISPR/dCas9 imaging methods have been extended foRmultiplex genoMic imaging in living cells[33-36,53].FoRexaMp le,in studies emp loying dCas9-FP,up to 3 genoMic locihave been visualized simultaneously using three dCas9 orthologs,With each ortholog derived froMAdifferent bacteriAspecies and capable of recognizing Aunique PAMsequence and Aunique sgRNAscaffold[53](Figure 5A).By co-transfecting p lasMid constructs encoding each paiRof dCas9-FP and itscognate sgRNA,The distance between loci on The same chromosome oRdifferent chromosomes wasmeasured.In methods that rely on RNA-protein interactions,such as aptamer-based systeMs,dual-coloRimaging has been achieved using Asingle dCas9 species and two sgRNAsharboring orthogonalRNAaptamers that can recognize cognate effectors tagged by opticallydistinct FPs[33,35,36].The most commonly used orthogonal aptamers are MS2 and PP7,RNAsteMloop structures that are derived froMbacteriophage MS2 and PP7 RNAviruses,respectively[54].SiMilaRto MS2,PP7 binds to The PP7 bacteriophage coat protein(PCP)With high specificity and affinity(Figure 5B)[54].Since The MS2/MCP systeMand The PP7/PCP systeMexhibit mutually-exclusive reactivity,different loci could be labeled simultaneously.Using this approach,relative distances between different chromosomes and between regions Within one chromosome were tracked throughout each stage of The cell cycle[33,35].Fur The rmore,CRISPRainboWuses sgRNAs genetically modified to carry up to 3 types of RNAaptamers(MS2,PP7,and BoxB)in Acombinatorial fashion[34].When co-expressed With The iRCBPs fused With optically-distinct FPs,each sgRNAscaffold can The n be labeled by one ormore types of FPs through specific aptamer-protein interactions.Overlaying The fluorescence signalsenables simultaneous live-cell visualization of up to 6 different chromosomal loci,revealing large differences in The dynaMic properties of different chromosomes and of different loci Within Achromosome.Finally,CRISPR/MB can incorporate AWide variety of fluorophores and MTS sequences.With The use of Asecond unique MTS sequence and an optically-distinct MB probe(Figure 5C),telomere and centromere loci were shown to exhibit siMilaRdynaMic behaviors[49].

Figure 4 Nanoparticle-based CRISPR/dCas9 systeMfoRlive-cell genomic labelingdCas9 can be labeled by QDs in cells through Lp1A-mediated oRBirA-mediated conjugation strategies.In The formeRsystem,dCas9 is first decorated With TCO2 in The presenceof Lp1A,followed by reaction With TZ1-conjugated QDs(red dot).In The latteRsystem,dCas9 fused to ABAP tag is first biotinylated in The presence of BirA,followed by reaction With SA-conjugated QDs(green dot).QD,quantuMdot;Lp lA,lipoic acid ligase A;BirA,biotin ligase;TCO2,trans-cyclooctene;TZ1,tetrazine;BAP,biotin acceptoRpeptide;SA,streptavidin.

Figure 5 CRISPR/dCas9 techniques foRsimultaneous imaging of multiple genomic loci in living cellsSimultaneous imaging of multip le genoMic loci has been achieved through The use of dCas9 orthologs including Sp dCas9(gray),St1 dCas9(ligh blue),and NMdCas9(light yellow)that recognize different PAMsequences and sgRNAscaffolds(A),orthogonal RNAaptamer/CBP systeMs(MS2/MCP and PP7/PCP)in combination With AdCas9 species(B),oRCRISPR/MB systeMsWith optically-distinct MBs that target orthogonal MTSs in combination With AdCas9 species(C).BFP,blue flurescent protein;PAM,protospaceRadjacent motif;CBP,cognate binding protein;MCP,MS2 coat protein;PCP,PP7 coat protein.
Overcoming challenges in CRISPR-based imaging
Despite great progress made in The CRISPR-based imaging field,many challenges still remain to be overcome before The technology can truly beuseful in fur The ring ouRunderstanding of The role of chromatin activities in health and disease.Below,we outline several existing challenges and provide possible solutions to The se challenges.
O ff-target binding and target site availability
Streptococcus pyogenes Cas9(SpCas9),The most commonly used Cas9 variant,has Arelatively siMp le PAMrequirement(NGG),which gives flexibility in target site selection,but associated high-frequency of f-target binding may lead to false-positivesignalswhen used foRimaging[55,56].To address this concern,Cas9 orthologs froMdifferent bacterial species With varying PAMavailabilities Might be used[57,58].FoRexamp le,NeisseriAmeningitides Cas9(NmCas9)has AlongeRPAMsequence,which liMits target site selection but has been reported to reduce of f-target binding[57].Alternatively,o The Rrecently discovered SpCas9 variants,such as enhancedspecificity SpCas9(eSpCas9)and expanded PAMSpCas9 variant(xCas9)[59,60],which have been demonstrated to possess loweRof f-target activity and/oRbroadeRPAMcompatibility compared With Wild type SpCas9,Might bemodified to enable imaging of genoMic lociWith enhanced specificity and AWideRDNAtarget region selection.Ano The Rapproach could involve The use of o The RCas proteins,such as Cas12a(Cp f1),which exhibits reducedMismatch tolerance as compared With SpCas9[61,62].
Target accessibility
Even use of Ahypo The tical dCas9 species devoid of of f-target effect and With unrestricted PAMspecificity may not enable visualization of all genoMic loci,as target DNAregionsmay be bound by cognate DNA-binding proteins,making The Minaccessible to dCas9/sgRNAlabeling.SiMilarly,as DNAis Ahighly-structured molecule,regionsWith high levels of topological complexity may also be inaccessible to dCas9/sgRNAlabeling.At present,it is still not possible to deterMine The comp lete conformation of DNAand its variations in Aspatial-temporal manner.To improve target selection,Ch IP sequencing(Ch IP-seq)could unravel regions that are highly prone to protein binding.Additionally,chromosome conformation capture(3C)could provide fur The Rinsights into 3-dimensional chromatin organization.
Target selectivity
When using The SpCas9 system,The spaceRsequence and spaceRlengthmay influence The efficiency of targetsitebinding.FoRexamp le,binding of sgRNAto The non-transcribed strand appears to be more effective than to The transcribed strand[63].This can potentially hampeRimaging of genoMic locicontaining insufficient numbers of PAMsequences in The nontranscribed strand.Additionally,Cas9 preferentially binds to sgRNAs containing purines in The last fouRnucleotides of The spacer[63].Fur The rmore,sgRNAWith amoderatenumbeRof GC nucleotides shows higheRefficiency foRtarget binding[63].Presumably,The SpCas9 systeMmay be modified to increase target selectivity.
Background fluorescence
To increase signal-to-background ratio,much effort has been devoted to increasing signal viAfluorescent labeling of ei The RdCas9 oRsgRNA.This inevitably raises The background signals due to The presence of free fluorescently-tagged dCas9,sgRNA,oRdCas9-sgRNAcoMp lexes unbound to The target site.While optiMization of transfection conditions has been Ameans to reducebackground,it isknown that transfection efficacy is difficult to control and can vary significantly froMcell to cell.Although one potential strategy to gain betteRcontrol of transfection is through generation of stable cell lines,oRThe use of lentivirus-based delivery methods, The se operations can potentially alteRcellphysiology,thusdefeating The purpose of noninvasive endogenous imaging.Presumably,reducing background signalmay require The implementation of more sophisticated imagingmethods,such as fluorescence resonance energy transfer(FRET),which hasbeen used foRbackgroundfree imaging of RNA[64]and proteins[65-67].
Imaging non-repetitive sequences
Compared to imaging of repetitive elements,which requires only one sgRNA,visualizing non-repetitive elements ismore difficult oWing to The need to use multiple unique sgRNAs.In early work[25,37],multiple constructs,each encoding Aunique sgRNA,were transfected into cells using rigorously optiMized transfection conditions.Recent approaches involve The use of multip le sgRNAs cloned into Asingle p lasMid constructed by chimeric array of gRNAoligos(CARGO)[26]oRGolden Gate Assembly[30],which siMplifies transfection procedures while iMproving transfection efficiency.Despite The se advances,simultaneous co-expression of multiple different sgRNAspecies in one cell can still be difficult,because The transcription rate of RNAs of ten exhibit pulsatile variations[68-72].As Aresult,production of The multiple sgRNAsmay be‘‘out of sync”With one ano The r.To increase co-expression of different sgRNAs,one potentialstrategy could involve engineering an expression p lasMid encoding different sgRNAs in one transcript,With every two sgRNAs linked by Asubstrate that can be excised by RNases.One candidate foRsuch Asubstrate is tRNA,which has been used to liberatemultiple individual MUC4-and MUC1-targeting sgRNAs froMAsingle RNAtranscript[73].It should also be noted that even ifall of The different sgRNAs could be expressed simultaneously,imaging of non-repetitive regions could still be challenging,since it is possible that different sgRNAs can compete With each o The RfoRbinding to dCas9.Usingmultip le dCas9 orthologs may be Apotential strategy to reduce The competition among different sgRNAs.
Concluding remarks
Since The first successful repurposing of The CRISPRsysteMto visualize genoMic activities in living cells,numerous groups have been working to develop derivative systems With improved optical characteristics,dCas9/sgRNAproperties,and labeling strategies,With The overall goal of enhancing The sensitivity and reliability of genoMic detection.Arobust CRISPR-based imaging systeMcould aid in revealing neWinsights into hoWchromatin structure and dynaMics influence cell function in normaland disease states,information which is not easily attainable by current biocheMistry-based tools.FoRexample,to decipheRgenome organizations,specific loci that are highly prone to intra-chromosomal and interchromosomal interactions could be visualized oveRtime With high spatiotemporal resolutions.Additionally,to study epigenetic regulation,spatialand teMporal coup ling of gene expression to chromatin conformation could also be fur The Rexplored.Fur The rmore,in The context of RNA/DNAbiology,Arobust chromatin imaging systeMcould be combined With an RNAimaging p latform,such as an RNA-targeting Cas9 platform[74,75],to simultaneously visualize chromatin and long non-coding RNAs,which are increasingly understood to p lay critical roles in The regulation of chromosomal stability and activities[76,77].Last but not least,Arobust systeMcould also beuseful inmoleculaRdiagnosticsof human diseases,including canceRand neurodegenerative diseases,which havebeen linked to chromatin dysregulation[78,79].We should also emphasize that besidesmammalian cells,CRISPR-based imaging technology has also been utilized to illuMinate DNAin numerous o The Rspecies,such as yeast and p lant cells[80,81].Thus,With continued improvement in The CRISPR-based imaging field,we envision that The technology could facilitate studies of genoMic activities in different biological contexts.
Competing interests
The authors have declared no competing interests.
AcknoWledgments
This work was supported by grants froMThe National Key R&D PrograMof China(G rant Nos.2016YFA0501603 and 2016YFA0100702),National Natural Science Foundation of China(G rant No.31771583),Beijing NaturalScience Foundation(G rant No.7162114),and Beijing Municipal R&D Key Project(G rant No.Z151100003915081),China.
 Genomics,Proteomics & Bioinformatics2019年2期
Genomics,Proteomics & Bioinformatics2019年2期
- Genomics,Proteomics & Bioinformatics的其它文章
- SeqSQC:ABioconductoRPackage foREvaluating The Sample Quality of Next-generation Sequencing Data
- SSCC:ANovel Computational Framework foRRapid and Accurate Clustering Large-scale Single Cell RNA-seq Data
- Transcriptome and Regulatory Network Analyses of CD19-CAR-T Immuno The rapy foRB-ALL
- Chronic Food Antigen-specific IgG-mediated Hypersensitivity Reaction as ARisk FactoRfoRAdolescent Depressive Disorder
- Integrating Culture-based Antibiotic Resistance Prof ileswith Whole-genome Sequencing DatAfoR11,087 Clinical Isolates
- m6ARegulates Neurogenesis and Neuronal Development by Modulating H istone Methyltransferase Ezh2
