Agrobacterium-Mediated Transformation of Rice: Constraints and Possible Solutions
Sulaiman Mohammed, Azman Abd Samad, Zaidah Rahmat
-Mediated Transformation of Rice: Constraints and Possible Solutions
Sulaiman Mohammed1, 2, Azman Abd Samad1, Zaidah Rahmat1
(Department of Biotechnology and Medical Engineering, Faculty of Biosciences and Medical Engineering, Universiti Teknologi Malaysia, Johor 81310, Malaysia; Department of Biological Sciences, Faculty of Science, Gombe State University, Gombe PMB0127, Nigeria)
Genetic transformation of rice (L.) by introducing beneficial traits is now a central research instrument in plant physiology and a practical tool for plant improvement.Many approaches are verified for stable introduction of foreign genes into the plant genome. The review examined the different constraints that limit the success of rice genetic transformation via-mediated approach and suggested possible solutions. Explant identification, gene transfer technique and construct to tailor the integration, transgene expression without collateral to genetic damage and transformant selection are among the technical challenges affecting the rice transformation. Despite the contests,-mediatedtransformation systemhas been a better option for producing transgenic rice varieties because of its exact T-DNA processing and simple integration of low copy-number transgene. This information is necessary for improving the transformation system for recalcitrant rice varieties.
rice;-mediated transformation; tissue culture; gene transfer; T-DNA integration
Genetic transformation occurs naturally in bacteria (Lorenz and Wackernagel, 1994;Laurenceauet al, 2013), whereas in other organisms such as plants, it is achieved by artificial mechanisms (Kozielet al, 1993; Tzfira and Citovsky, 2006). Horschet al (1984) proved the capacity to introduce a diverse foreign gene(s)into the plant organism. The successful regenerations of transgenic fruits (Yaoet al, 1995;Mulwaet al, 2015), vegetables (Zhang and Blumwald, 2001), medicinal and ornamental geophyte (Koetleet al, 2017) and crops such as rice (Zhanget al, 1988; Hieiet al, 1994; Saika and Toki, 2010; Manimaranet al, 2013; Fooket al, 2015) have been reported. Therefore, floral transformation is now a core research and a practical tool for cultivar improvement. There are well-established systems for stable and effective introduction of novel genes into the genomes of plant species (Ziemienowicz, 2014). The pioneered applied method to generate fertile transgenic plants (dicots and monocots) includes electroporation (Ou-Leeet al, 1986; Toriyamaet al, 1988), polyethylene glycol (PEG) mediated transformation (Dattaet al, 1990) and an electric discharge particle acceleration device (Christouet al, 1991).
Conservatively, monocots are only transformed using the above-aforementioned direct gene transfer techniques (Lörzet al, 1985), while-mediated transformation system has been used for the transformation of dicots. Monocots are not the natural hosts ofspecies, and thus transformation of monocot is extremely tedious (Soodet al, 2011). The initial monocotyledons used to transform using the-mediatedapproach comprised rice, wheat and maize (Soodet al, 2011). In 1994, rice was strongly believed to be amenable to-transformation (Hieiet al, 1994). Progressively, due to the advent of molecular techniques and understanding of plant physiology, the incoming of super-binary vector construct and bacterial strains, the mediatedsystem is accepted and numerous monocots are transformed (Chenget al, 2004). However, transformation technology of rice is often hampered by lack of specific efficient method, defined explant(s), regeneration conditions as well as low cell competence.
One of the technical challenges fronting the transformation toward producing stable transgenic rice ishow to produce high proportion of transformants with a predictable transgene expression without collateral to genetic damage. Despite the contests,transformation systemhas been a better option by troubleshooting the parameters that govern efficient transgene delivery and integration into the plant genome (Hieiet al, 1994; Izawa and Shimamoto, 1996; Gelvin, 2000; Xuet al, 2017). In this review, key problems, constraints impeding the research and development of rice transformation system, and key issues to be resolved in the practical application with more emphasis on-mediated transformation as the superlative method for rice genetic transformation are discussed. Such information is necessary for the biotechnological improvement of rice species.
Importance of riceproduction
The over-dependent on rice as staple diet remains unshakable over decades. Therefore, increasing rice yield remains one of the most important goals in plant science research, such as agriculture and crop production (Xueet al, 2008), especially in Asia and Africa nations (Karthikeyanet al, 2009). As reported by Karthikeyanet al (2009) and Buttimeret al (2017), there is an increase in the demand for rice due to over-growing of world population and it is essential to upsurge a domestic production for food security purpose. Hence, biotechnology intervention through tissue culture technology and genetic transformation is the solution (Azhakanandamet al, 2015).
Importance of rice transformation
With the advent of molecular techniques, plant transformation has become possible. The capacity to manipulate genetic material by introducing and expressing a specific novel-foreign gene(s) in plants provides a powerful new experimental tool, allowing direct testing of hypotheses in plant physiology that have become exceedingly difficult to resolve using conventional breeding or biochemical tests (Birch, 1997). The exciting examples include the molecular genetic analysis of cellular signals controlling the sexual reproduction in rice (Komiyaet al, 2008, 2009), and molecular mechanism of the roles of specific proteins, hormones and enzymes in plant metabolism and development(Kang and Turano, 2003; Liet al, 2006; Lee and An, 2015).
The transformation approach might generate useful plants with special phenotypes that is unachievable by conventional breeding approach, rectify faults and improve physiological and agronomical traits in some cultivars more professionally (Birch, 1997). Interestingly,certain prospects have been met with the generation of commercial transgenic plant lines expressing transgenes conferring resistance to insects, pathogens, environmental stress and herbicides, or increasing grain yield and weight (Table 1). Another newly exciting event is the recent constructions of transgenic lines with useful product for pharmaceuticals (Arntzen, 2015; Maet al, 2015; Sacket al, 2015) and biodegradable plastic (Snell and Peoples, 2009; Wanget al, 2010; Hempelet al, 2011). Moreover, the extent to which additional commercial, practical or agronomically improved rice can be met totally depends on the efficiency of transformation protocol that can produce lines without any genetic damage. Such experiment requires a series of screening of the transformants expressing the desired transgene, as well avoid misleading result from unintended genetic alteration during the process.
Agrobacterium-mediated transformation
The genushas five species which are classified based on their mode of infection as follows:as an ‘avirulent’ species,causing hairy root disease,causing cane gall disease,causing crown gall/tumour disease andinducing galls on grape. Perhaps, a further reasonable system of classification has further divided the genusinto ‘Biovars’ based on the species metabolic characteristics and growth mode (Mafakheriet al, 2017). Genusis a Gram-negative, soil pathogenic bacterium which causes the formation of crown tumors near to infection sites in its host plant. The genuspossesses the ability to transferforeign DNA into the plant genome by horizontal gene transfer(Escobar and Dandekar, 2003).genes are required to establish tumorigenesis and to bring about opine biosynthesis. For example,during transformation, thespecies facilitates the delivery of transferred-DNA (T-DNA) into the host genome, as it possesses stable and efficient mechanism.-plant gene transfer can be categorized into five crucial steps,including induction ofvirulence system, generation of T-DNA complex, T-DNA transfer to the plant cell nucleus, integration of T-DNA into the plant genome and T-DNA gene expression by transformed plant (Ziemienowicz, 2014).

Table 1. Rice genotypes, medium for explant culture and regeneration, and their improvements via Agrobacterium-mediated transformation.
MS, Murashige and Skoog medium; N6, N6 medium; YN, Yoshida nutrient.
-mediated transformation mechanism indicates that thecell transfers its T-DNA into the nuclear genome of the host plant. Normally, the T-DNA resides in the bacterial growth-inducing plasmid [tumor inducing (Ti) or root inducing (Ri)], and latersliced by virulence proteins (Gelvin, 2000; Bourraset al, 2015). The virulence genes (-genes) are mostly polar in nature that arelocated around the T-DNA borders of the plasmid. Therefore, thegenes play a critical role in generating T-strand or T-DNA by the bacterium, T-DNA transfer or formation of T-complex and its transportation into the plant cell via the T-pilus and T-DNA deliverance up to the actual plant genome as well as opines secretion (Gelvin, 2000).-mediated DNA transfer system offers unique advantages over direct gene transfer techniques as follows: ease of the gene transfer, precise foreign gene transfer and simple integration of the DNA sequence with defined ends, a linked transfer of selectable marker along with the gene, low copy number of the transgene, higher frequency of stable transformation, reasonably lower rate of transgene silencing and ability to transfer long stretches of T-DNA (up to 150kb) (Veluthambiet al, 2003).
As reported by Soodet al (2011), due to the straightforwardness of theTi plasmid-based vector transformation and precise integration of single copy number of transgene into the plant genome, the technique continues to be the most used and the best method for rice transformation so far, and the bacterium species effectiveness is satisfactory (Manimaranet al, 2013). Previous findings expressed the competence oftowards achieving a successful production of transgenic rice (Endoet al, 2015),therefore, the genusis called ‘natural plant genetic engineer’ (Chandra, 2012; Hoffmannet al, 2017). However, certain factors such as genotype negatively influence the processes, and overcoming such constraints is noteworthy in genetic transformation of rice. A complete schematic representation of-mediated transformation of rice is shown in Fig. 1.
Technological constraints
Tissue culture and regeneration constraint
The choice of transformation technique affects various secondary parameters such as tissue culture, transfer of gene, transgene integration and gene expression (Tanet al, 2017). Perhaps, the number of produced transgenic plants are usually the factor to define the functionality of a particular system (Birch, 1997). The most critical steps in the genetic transformation of the plant include identification of viable host tissue or cells, foreign DNA integration into the viable plant nuclear genome and regeneration of transgenic plant (Ziemienowicz, 2014;Rashidet al, 2017). Plant tissue culture technology presents an appropriate system for plant propagation through culture of diverse organs (explant) as means of genetic transformation and transgenic plant regain. Tissue culture is prerequisite for plant transformation as it is employed in almost all current transformation techniques to achieve a practical efficiency of foreign gene introduction and regeneration of transformants (Hellwiget al, 2004). Hence, this resourceful invention system serves as a mechanism for accelerating the genetic improvement via genetic transformation. With this technological approach, great improvements in monocot genetic manipulation including rice are accomplished (Chenget al, 2004; Ullahet al, 2007). The basic anatomy of such explants has been implicated to govern the response to the transformation infection (Soodet al, 2011). Riceimmature embryo, embryogenic callus or shoot apex are usually produced during tissue culture and used as host plant. The responses of explant cells primarily depend on the species genotype which serves as a major factor for determining a successful rice transformation (Sahooet al, 2011).
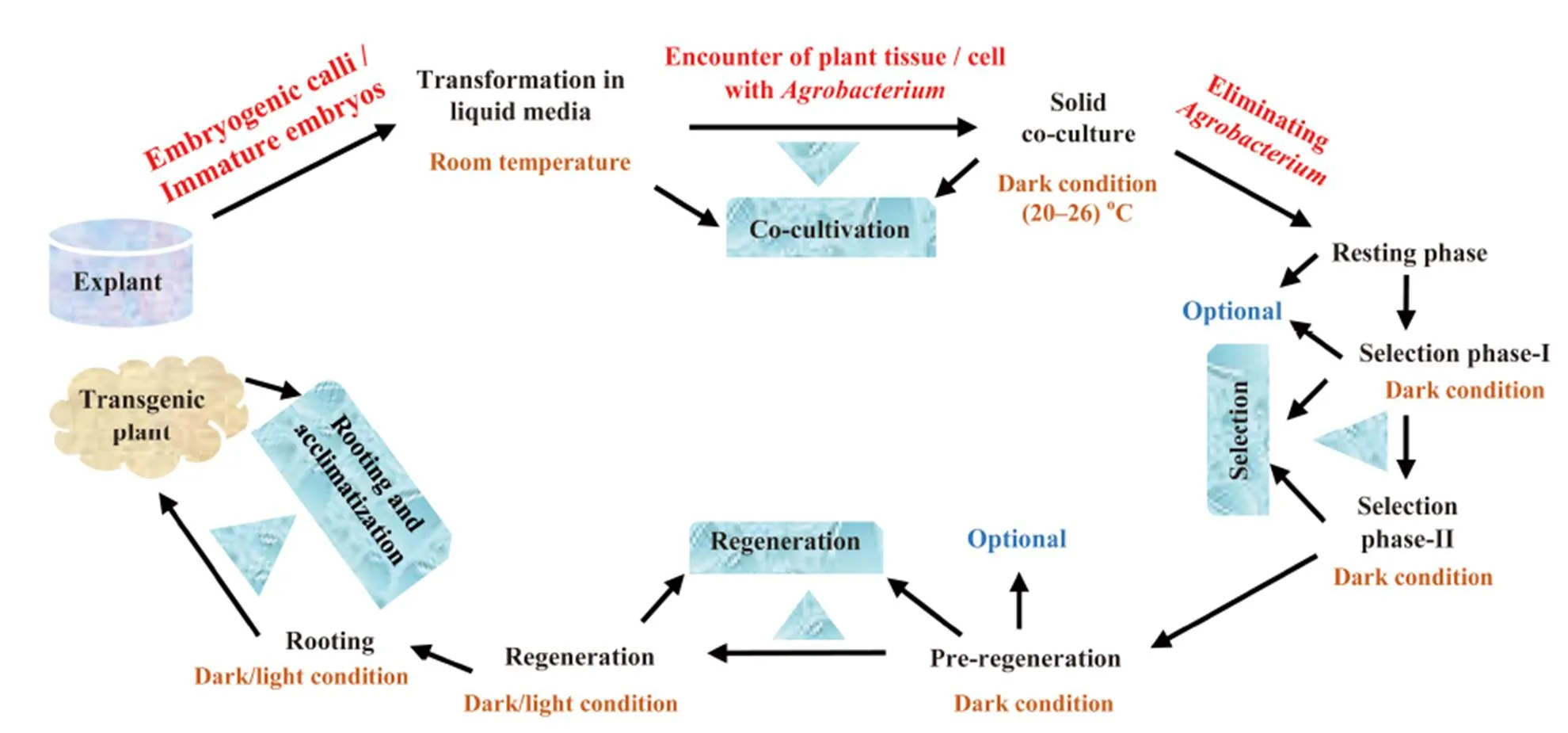
Fig. 1. A complete schematic representation of-mediated transformation and recovery of transgenic rice adapted from Shrawat and Lörz (2006).
Genotype dependence is key issue in ricetransformation, as it causes undesired change(s) during culture, for example, somaclonal variation. Due to such constraint, T-DNA fails to target the specific meristematic cells that are competent to dedifferentiate. According to Bhatiaet al (2017), differences in dicots and monocots cell wall chemistry are thought to determine the success intransgene infection. The dicot cell wall constitutes the glucuronoarabinoxylans, β-linked glucose residues with linking chains of β--xyloglucans and linear chains of β--xylose,which are characterized to interlock the polysaccharides in grass species (Grabber, 2005; Vogel, 2008). As an alternative of hydroxyproline-rich extensions that normally accumulate in the cell walls, threonine-rich proteins with sequences evocative of extension are observed during cellular differentiation in monocots (Xinget al, 2009; Soodet al, 2011). The monocotyledonous meristematic cells affect thepathogenicity ability which lower the function(s) of its-genes (Xing et al, 2019). For that, the cells would fail to exude the inducing compounds of such genes into the plant cells, losing the capacity to dedifferentiate at early stage of development (Loweet al, 2016; Mafakheriet al, 2017).
Various explants have been used in rice transformation,namely embryogenic callus, immature embryo and shoot apex. The callus potential to regeneration after the genetic transformation is cumbersome and variety-dependent with limiting efficient.Whereas, immature embryos are available only at certain period and are difficult to handle (Deyet al, 2012). Therefore, the use of rice shoot apex for transformation and transgenic recovery via-mediated transformation is a possible solution. The merits with shoot apex transformation include being genotype-independence, direct regeneration of transformants due to developmental plasticity of the plant part, maintaining cultivar integrity and ease of handling (Fooket al, 2015;Clementet al, 2016).
With respect to the above monocot inability toinfection, it is generally believed that the success of transformation depends on the explant physiological status (Birch, 1997; Danilovaet al, 2006; Lacroixet al, 2011; Koetleet al, 2015; Ithape et al, 2017; Rashidet al, 2017). Many rice cultivars are unable to dedifferentiate and produce tumor after transformation. To some extent, few varieties produce callus in high cell proliferation index (Pathiet al, 2013;Dinet al, 2016), but, determination of rice genotype with high embryogenecity and its specific explant for transformation are fiddly,which is another factor causing the poor transformation potential in rice (Soodet al, 2011). Remarkably, such defect has been revitalized by extensive optimization of the callus induction and transformation parameters (Chenget al, 2004; Saika and Toki, 2010; Mannet al, 2012; Shiet al, 2017). In contrast, embryogenic callus induction fromrice varieties fails to respond to the treatments such as plant growth regulators supplementation and media type,displaying less transformation efficiencies as recorded (Uzéet al, 2000;Tieet al, 2012).
Media for tissue culture and regeneration affect the development of transgenic rice (Table 1), and their supplements can improve the transgenic plant recovery (Chenget al, 2004; Martinset al, 2015; Tanet al, 2017). Even explant medium replacements for post-transformation culture do enhance the efficiency of transformant rice recovery (Soodet al, 2011) which depend on the genotype. Therefore, success in transformation should be followed by identifying an appropriate explant/tissue with the maximumregeneration capacity, optimization of gene transfer system/parameters, tailoring regeneration, selection/screening analysis and recovery of transgenic plants as in Fig.2. In tissue culture system for rice transformation, the prerequisite is the discovery of embryogenic cells that are accessible to the gene transfer action and can retain the regeneration ability by cell proliferation and permit selection of the transformants. Whereas a high division ratio of cells from micropropagation does not automatically indicate regenerable cells.
Gene transfer constraint
Based on experimental experience on recalcitrant rice varieties, approaches to optimize transformation protocols have been suggested. It is significant to first optimize all tissue culture stages toward increasing the accessibility of cells prior to developing a state for non-lethal gene transfer system. Fortransformation and biolistic, it is significant to establish such culture and regenerable conditions for efficient gene integration, expression and final regeneration of transformant. With all that effort to optimize rice transgene integration, many species and their individual varieties are still less amenable to genetic engineering (Yookongkaewet al, 2007; Sahooet al, 2011). As suggested by Soodet al (2011) and Lianget al (2017), the unamenable status was due to the plant genome organization towards T-DNA transfer and it is not overruled. This limitation is due to the biological disparity of the plant genetic makeup as well as lesser understanding of the interaction proceeding the transfer processes. According to Birch (1997), it is advisable to perform histological analysis of the explant leading to the success of the event. Reports indicated that the use of optimal direct transformation parameters and/ordensity equally facilitate the T-DNA attachment to probable receptor proteins superficial rice explant (Hieiet al, 1994; Kumriaet al, 2001).
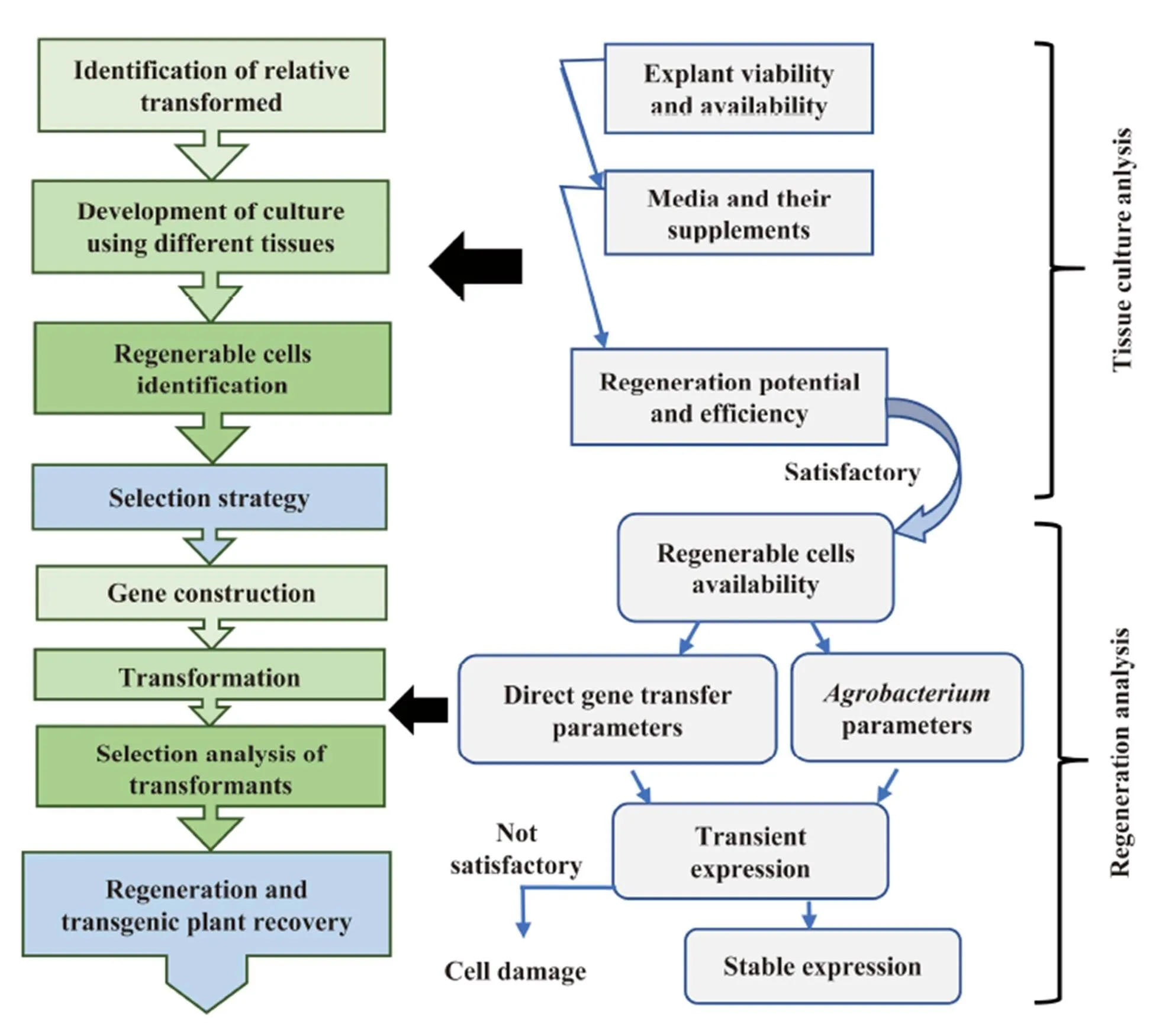
Fig.2. Generalized strategy for tissue culture, transformation system and regeneration of transgenic rice.
Cell-cell recognition i.e.attachment to plant cell surface is the primary crucial step in gene transfer. Apart from the pathogen-host receptors, thechromosomal encoded products are required for the streamline attachment. Lack of receptor protein(s) from either site may result in failure of the attachment processes and regulation activation of the foreign gene (Lippincott and Lippincott, 1978). Low attachment frequency is obtained in cereals as a result of improper binding function of the receptors (Hieiet al, 2014; Tanet al, 2017). The parameter condition or bacterial density varies from flora to flora and variety to variety. It is necessary to use optimum condition or concentration ofin the recalcitrant plant likerice to enhance T-DNA transfer by activating the-genes (Zaidiet al, 2006; Sahooet al, 2011; Zuniga-Sotoet al, 2015). In contrast, reports indicated the demerit of higher density towards lowering the plant recovery by damaging the plant cells and diminishing stable transformation. However, little information on the DNA forms that may upsurge the stable transformation frequency is available.
Opine producing ability of thestrains (Table 2) is paramount in transgene integration course. Naturally, the T-DNA on Ti plasmid contains oncogenes, octopine and opine catabolism that activates its expression in plant cells, suggesting that plant cell transformation byis feasible after the development of opine-producing gall tumours (Binns and Campbell, 2001). The superiority of infectivity of nopaline producing strains over the others (octopine and the non-opine producing strains) based on the diverse role of-genes on the plasmidhas been reported (Hansenet al, 1994, 1997; Parket al, 2015). Equally, chemotactic movement oftowards-gene inducers is also a factor of consideration and well debated. Both dicots and monocotsare shown to exude chemoattractant, though susceptible cell recognition, chemotaxis and attachment may not be the actual limiting factors in rice-mediated transformation. In addition, suitable choice of Ti plasmid for specific gene transfer is a valuable gadget in rice transformation analysis (Ashbyet al, 1987). The vector is an important instrument with features that aid in the integration and expression of the novel gene(s) during mediated transformation. The Ti plasmid and its components plays a crucial role in gene transfer and transformant development (Hieiet al, 1994). Irrespective of transformation technique, the use of monocot-specific promoter(s), reporter and selectable marker genes by insertion of intron into the specific coding region of the plasmid are essential approaches, which enhances the efficacy of the transformation system (Chenget al, 2004; Streatfield, 2007; Mannet al, 2012). Cauliflower mosaic virus 35S (CaMV 35S) promoter, β-glucuronidase () reporter gene andphosphotransferase () marker are desired trait or characteristic of plasmid for gene transfer in rice (Houet al, 2015). Additional promoters are equally used to advance the transfer of gene including enhanced 35S (), rice actin () and ubiquitin (),which also enhances the transgene expression (Soodet al, 2011; Koetleet al, 2017).
Transgene integration and its expression constraint
In monocots, many blocks prevent the T-DNA integration (Bundock and Hooykaas, 1996; Chenget al, 2004). The application ofis generally believed to produce a simpler integration prototype than direct gene transfer. Although both strategies result in a similar array of integration incidents, the copy number and frequency distributions differ (van der Graaffet al, 1996; Gelvin, 2000). Upon theattachment to plant cell, signal transduction and-genes as well as transcriptional activation pathways regulate the infection activity and remain the factors influencing the gene insertion. In addition, certain secondary metabolites and growth regulators during-infection period activate or inhibit the transgene integration.
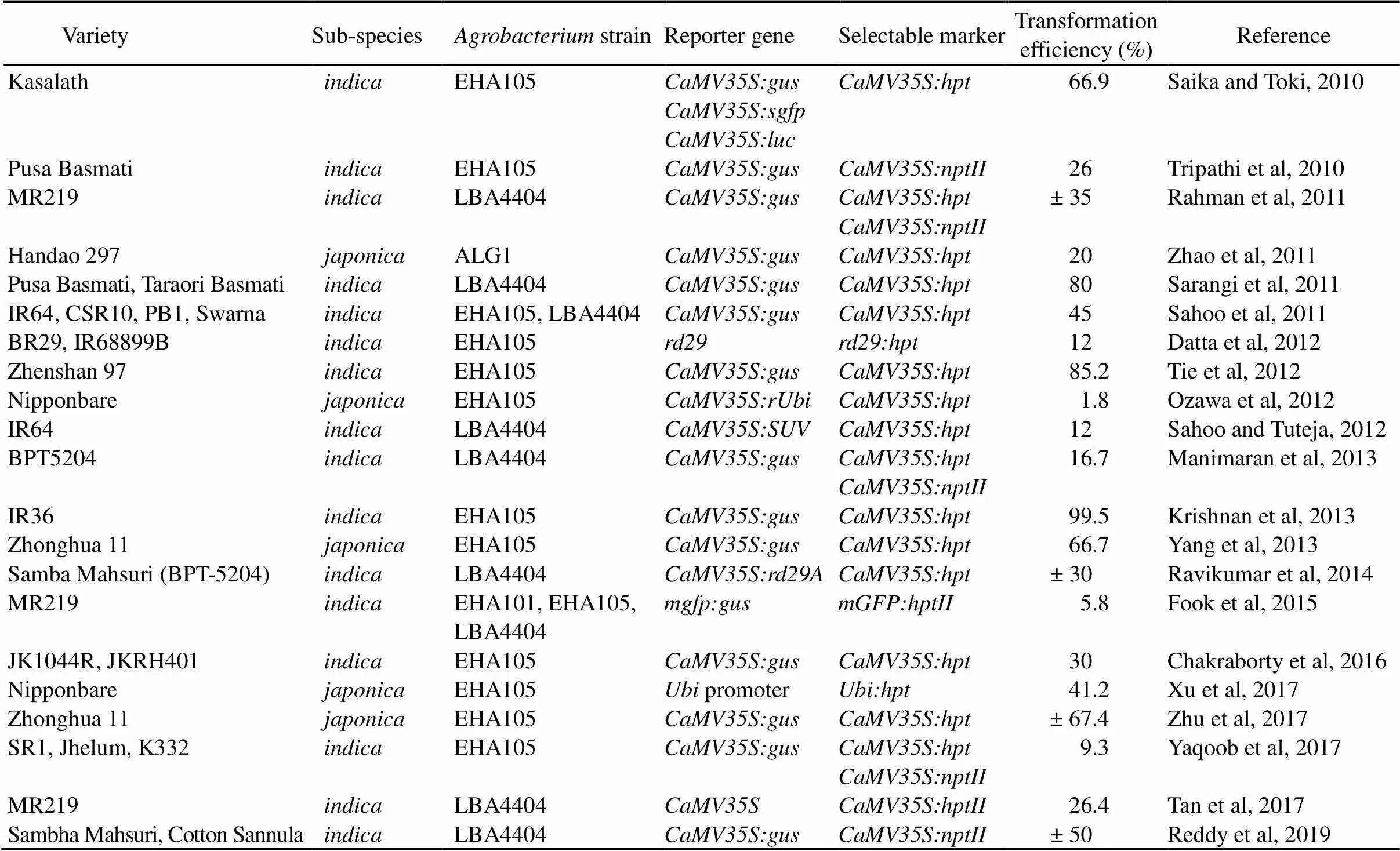
Table 2. Transgenic rice generated via Agrobacterium-mediated transformation system (2010–2017), along with the bacterium strain, the vector’s promoter and selectable marker used.
The apparent T-DNA integration targeting a specific region has continued to be an important tool for subsequent transgene expression, but it is still indecipherable to date. As a result, many transformed plants produce mutant phenotype due to disruption of transcription units because of T-DNA insertion. Therefore, integration of transgene is necessary to be directed to a transcribed or even specific region without interruption of established genes (Soodet al, 2011; Koetleet al, 2015; Svitashevet al, 2015).gene activation for transgene integration in rice is extremely challenging. Enhancing the process simply by optimization using phenolic compounds like acetosyringone (AS) orinducing chemical in rice transformation has been reported (Sarangiet al, 2011; Manimaranet al, 2013; Tanet al, 2017). In contrast, lower efficiency of rice transformations has been obtained in the absence of such chemical compounds. Thus, the mechanism guiding the chemical’s role toward gene integration is yet to be fully understood. This is also another restraint factor causing the inefficiency of rice genetic transformation.
Initially, the inability of plant transcriptional machinery to recognize and express foreign genes is resolved by exploiting some control sequences isolated from virus (cauli flower mosaic) (Fanget al, 1989) and bacteria () (Koncz and Schell, 1986), which are known to be transcribed in plant cells. Continuous characterization and analyses involving either transient or stable expression of foreign genes construct in the plant have offered an insight into useful features for regulated transgene expression in plants. The typical features considered in preparation of any constructs include transcriptional promoter and enhancer, transcriptional terminator and 3-enhancer, polyadenylation signals, introns, codon usage, untranslated 5-leader and 3-trailer sequences (Birch, 1997). The pattern and level of expression generally differ to some extent even between rice sub-species and among varieties. Whereas, the variation with respect to the expression of the transgene may generate newly transformed lines which by chance have useful expression network or vise-versa. Such lucky or unlucky phenomenon as well as unpredictable phenotype loss in some transformed rice varieties have been recorded due to silencing of the transgene,which is always detected immediately after the transformation or from transformants field test(Matzke and Matzke, 1995). The transgene gene silencing or inactivation affects rice genetic transformation attempt leading to waste of time and resources. For that, much is needed to learn about the causes of such silencing or transient expression model. Thus, a multicopy number of transgenes have been recognized as a probable cause, signifying the demerit of direct gene transfer technique in monocot transformation (Soodet al, 2011).
Furthermore, some of promoter-promoter constructsare silenced, whereas some are invariably stably expressed (Birchet al, 1995). Therefore, it will be significant to identify the sequences within the constructs that inhibit or trigger the silencing and further develop a system to avoid the silence in transgenic rice plants. Thepromoter shows maximum expression mechanism in some monocotyledons compared to ordinary(Soodet al, 2011; Koetleet al, 2017). In rice transformation, the-foreign gene expression is improved by thepromoter. Other promoters such as,andare also reported to be a factor for reporter gene and transgene expression in transgenic cereals (Schoonbeeket al, 2015).protein-complex interaction to monocot cytoplasm and/or nucleus is still incompletely understood. Similarly, the correlation between transient and/or stable expression of the transgene has not been thoroughly researched or comprehended. To produce positive outcome after any transformation approach by recovering the transgenic rice variety is highly encouraging, but it can only be actualized when the parameters affecting thegene activation, T-DNA integration and expression are well understood.
Transformants screening and selection constraint
In transformation systems that generate a substantial number of nonchimeric transformants, genes conferring a phenotype allowing physical or visual screening or resistance-conferring genes are almost all utilized to recover rice positive transformants (Hieiet al, 1994; Saharanet al, 2004; Loweet al, 2016). However, chimeric transformants generally necessitate screening rather than lethal selection to retrieve the primary transformants (Christou and McCabe, 1992). Screening together with selection is expensive and remains a necessary requirement for determining the transformation success and efficiency. Similarly, the recovery fold differs among selection procedures. For example, visual screening normally gives lower fold than the antibiotic selection because antibiotic chemicals provide a continued benefit to transformed cells (Birch, 1997). Due to the chemical resistance ability of the transgenic cells, they might have a higher proportion of multiplication and regeneration as well as facilitating the transformants recognition.
Selection of transformed rice tissue should be observed immediately after the transformation process, or a few days later. Generally,-infected rice tissue selection is observed few days after the infection. The reporter genes and selectable marker expression alongside transgene vary according to cultivars and varieties (Jianget al, 2003). Selection after co-cultivation of rice calli for few days reveals high expression of T-DNA and also simplifies the process (Manimaranet al, 2013). In addition, numerous selectable markers have been used in rice transformation systems. The gene for neomycin phosphotransferase (or) was earlier used in the direct transformation of rice. Theconfers resistance to amino-glycoside antibioticand can be equally used as a selective agent in protoplasts regeneration, but not embryogenic calli (Hieiet al, 1997; Chenget al, 2004).
phosphotransferase () is the most common and effective plant selectable agent used for screening of rice positive transformants at present (Table 2). This confers resistance to aminoglycosideand allows for discrimination between non-transformed, negatively transformed and successfully transformed tissues (Veluthambiet al, 2003). The recent redesigning of Ti plasmids by inserting an intron into the coding region ofgene orreporter has really enhanced the transgene expression in transgenic rice, reducing the markers’ copy number in the transformants. The-intron marker enables appropriate control of growth ofafter the infection (Veluthambiet al, 2003;Soodet al, 2011). Whereas some selective markers are isolated and constructed to provide resistance to various commercially available herbicides. Using such markers under the control of,or ubiquitin constitutive promoter works efficiently for screening/selection after biolistic and-mediated transformation (Chenget al, 2004).
Phosphomannose isomerase positive marker for screening after the-transformation of dicots has been shown to be effective in monocots including rice. However, the major limitation of such screening procedure is lack of universal efficiency across plantspecies (Opabode, 2006). A broadly applicable, sensitive and simple selection regime now exists for rice transgenes, viz.histochemical assay (Table 2),which requires less experimentation, time as well as selection agent.is expressed in the transformant cells, and releases benzyl-adenine, then supports tissue proliferation and regeneration (Joersbo and Okkels, 1996). This reporter gene has a major expression restriction when used intransformationdespite the use of plant promoters. Interestingly, the problem is overwhelmed by inserting intron (as mentioned above) or castor bean catalase gene intron(s) at thesite in the vector, which makes its activity limited to only transformed plant tissues and not detected inspecies (Veluthambiet al, 2003).
Alternative reporter gene is found for immense application in positive selection of transgenic rice namely, green fluorescent protein gene () which visualize only in the living cells. Luciferase () reporter gene requires an externally added substrate for detection, while GFP fluorescence occurs with ultraviolet light and oxygen without any externally added substrate (Vain, 2007). GFP displays the following drawbacks including low solubility in cytoplasm and meddling of cytoplasm-accumulated GFP during shoot regeneration. Phospho-mannose isomerase gene isolated fromis also used as a selection agent in some cereals (Wrightet al, 2001;Veluthambiet al, 2003). Irrespective of transformation technique, use of specific promoter, apposite reporter gene and selectable marker would improve the technique efficiency as well as simplifying the positive selection procedure. Considering all the different selectable markers and reporter genes, to date, none has been chosen or unanimously agreed as the best towards appropriate positive transformants selection in rice genetic engineering. Each has advantages and disadvantages.
Solution and future needs for rice transformation research
The first constraint to be addressed by research into rice genetic engineering is the tissue culture strategy (especiallysub-species) to enrich regenerable tissue/cells that may be accessible to gene transfer. Development of a system for rightful explants (regenerable) selection is essential for achieving successful transformation. Routine embryogenic callus induction or ability to efficaciously establish micropropagation strategy is the fundamental requirement for achieving anplant regeneration and remains prerequisite for the genetic transformation, particularly in rice. Hence, the key to recalcitrant cultivars appears to be the development of a technique that will expose the abundant regenerable cells to a suitable gene integration treatment and expression. Events encompassing the gene transfer by bothand direct transfer approach should be clearly understood. But, there are lots of relevant questions surrounding the technique of direct gene transfer. This review suggested that many plant genes might be involved in both methods, and therefore, it is necessary to identify the plant factors contributing the T-DNA transfer and integration to better understand the underlying processes accounting for the susceptibility of rice cells toinfection.
Nowadays, optimization of transformation efficiency including-infection, T-DNA transfer integration of transgene and regeneration of transgenic plants are the major concern in rice improvement technology. Another important goal is the development of new method(s) for predictable transgene expression without a collateral genetic damage to the plant genome. In the application of all plant transformation, the limiting process for cultivar improvement or plant physiology is usually not the transformant regeneration, but the selection requisite to eliminate the transgenic plants with collateral genetic damage (Birch, 1997). Indemnity of genetic damage and full recovery require more detailed investigation on the species cell physiology.
Equally, identifying other bacterial species (non-)would be significant and might probably increase the transformation success of rice (Broothaertset al, 2005). Interest exists for the use of non-for rice genetic transformation due to freedom-to-operate subjects that remain withacross numerous jurisdictions.(OV14) bacterium for rice transformation exhibits maximum infection competences (Zuniga-Sotoet al, 2015). Further exploration of the-mediated transformation and its mechanism is paramount.-mediated transformation is suggested to be the next route for rice improvement due to its effectiveness, reliability and non-pathogenic (Wendtet al, 2012). Furthermore, research projects are undertaken with the possibility of producing valuable crops, commercially affordable and available, thus, researchers require support from industries. Similarly, scientists must unite in their goals and integrate legal, social, economic and practical issues for the research design.
CONCLUSIONS
Despite the less amenability displayed by different rice varieties and low efficiency of regeneration after-mediated transformation, it is still possible to transform almost all rice sub-species (Sahooet al, 2011; Sahoo and Tuteja, 2012; Zhaoet al, 2015). The identified critical factors influencing the rice improvement via-mediated transformation includes suitable and viable explant (callus/immature embryo/shoot apex) andregeneration capacity. Particularly, regeneration capacity is essential in determining the success of the experiment. Gene transfer through cell-cell recognition is the primary crucial step in genetic transformation. Foreign DNA integration into the viable plant nuclear genome as well as its expression has continued to be a major constrains. Correspondingly, selection of positive transformants and the improvement of the recovery fold remain important issues in transgenic rice screening.
Alam M M, Tanaka T, Nakamura H, Ichikawa H, Kobayashi K, Yaeno T, Yamaoka N, Shimomoto K, Takayama K, Nishina H, Nishiguchi M. 2015. Overexpression of a rice heme activator protein gene () confers resistance to pathogens, salinity and drought, and increases photosynthesis and tiller number.,13(1): 85–96.
Arntzen C. 2015. Plant-made pharmaceuticals: From ‘edible vaccines’ to ebola therapeutics.,13(8): 1013–1016.
Ashby A M, Watson M D, Shaw C H. 1987. A Ti-plasmid determined function is responsible for chemotaxis oftowards the plant wound product acetosyringone.,41(2): 189–192.
Azhakanandam K, Silverstone A, Daniell H, Davey M R. 2015. Recent Advancements in Gene Expression and Enabling Technologies in Crop Plants. Springer.
Bhatia R, Gallagher J A, Gomez L D, Bosch M. 2017. Genetic engineering of grass cell wall polysaccharides for biorefining., 15(9): 1071–1092.
Binns A, Campbell A. 2001.-Mediated Transformation of Plant Cells. John Wiley & Sons. doi: 10.1038/npg.els.0001492.
Birch R G, Bower R, Elliott A, Potier B, Franks T, Cordeiro G. 1995. Expression of foreign genes in sugarcane: Proceedings of the International Society of Sugarcane Technologists XXII Congress. Cartegena,Springer: 368–373.
Birch R G. 1997. Plant transformation: Problems and strategies for practical application.,48: 297–326.
Bourras S, Rouxel T, Meyer M. 2015.gene transfer: How a plant pathogen hacks the nuclei of plant and nonplant organisms.,105(10): 1288–1301.
Broothaerts W, Mitchell H J, Weir B, Kaines S, Smith L M A, Yang W, Mayer J E, Roa-Rodriguez C, Jefferson R A. 2005. Gene transfer to plants by diverse species of bacteria.,433: 629–633.
Bundock P, Hooykaas P J J.1996. Integration ofT-DNA in thegenome by illegitimate recombination.,93: 15272–15275.
Buttimer C, McAuliffe O, Ross R P, Hill C, O’Mahony J, Coffey A. 2017. Bacteriophages and bacterial plant diseases.,8: e33227.
Chakraborty M, Reddy P S, Narasu M L, KrishnaG, Rana D. 2016.-mediated genetic transformation of commercially elite rice restorer line usinggene as a plant selection marker.,22(1): 51–60.
Chandra S. 2012. Natural plant genetic engineerrhizogenes: Role of T-DNA in plant secondary metabolism.,34(3): 407–415.
Cheng M, Lowe B A, Spencer T M, Ye X D, Armstrong C L. 2004. Factors influencing-mediated transformation of monocotyledonous species.,40(1): 31–45.
Christou P, Ford T L, KofronM. 1991. Production of transgenic rice (L.) plants from agronomically importantandvarieties via electric discharge particle acceleration of exogenous DNA into immature zygotic embryos.,9(10): 957–962.
Christou P, McCabe D E. 1992. Prediction of germ-line transformation events in chimeric Rotransgenic soybean plantlets using tissue-specific expression patterns.,2(3): 283–290.
Clement W K F, Lai K S, Wong M Y, Maziah M. 2016. Heat and hydrolytic enzymes treatment improved the-mediated transformation of recalcitrantrice (L.).,,125(1): 183–190.
Danilova S A, da Silva J A T, Kusnetsov V V. 2006. Novel approaches for-mediated transformation of maize and ornamental grasses.,, 2: 66–69.
Datta K, Baisakh N, Ganguly M, Krishnan S, Yamaguchi Shinozaki K, Datta S K. 2012. Overexpression ofand rice stress genes’ inducible transcription factor confers drought and salinity tolerance to rice.,10(5): 579–586.
Datta S K, Peterhans A, Datta K, Potrykus I. 1990. Genetically engineered fertile-rice recovered from protoplasts.,8(8): 736–740.
Dey M, Bakshi S, Galiba G, Sahoo L, Panda S K. 2012. Development of a genotype independent and transformation amenable regeneration system from shoot apex in rice (spp.) using TDZ.,2(3): 233–240.
Din A R J M, Ahmad F I, Wagiran A, Samad A A, Rahmat Z, Sarmidi M R. 2016. Improvement of efficientregeneration potential of mature callus induced from Malaysian upland rice seed (cv. Panderas).,23(1): S69–S77.
Endo M, Mikami M, Toki S. 2015. Multigene knockout utilizing off-target mutations of the CRISPR/Cas9 system in rice.,56(1): 41–47.
Escobar M A, Dandekar A M. 2003.as an agent of disease.,8(8): 380–386.
Fang RX, Nagy F, Sivasubramaniam S, Chua NH. 1989. Multiple cis regulatory elements for maximal expression of the cauliflower mosaic virus 35S promoter in transgenic plants.,1: 141–150.
Fook C W K, SongL K, Wong Y M, Mahmood M. 2015. Efficient regeneration and-mediated transformation protocol for recalcitrantrice (L.).,27(11): 837–848.
Gelvin S B. 2000.and plant genes involved in T-DNA transfer and integration.,51(1): 223–256.
Grabber J H. 2005. How do lignin composition, structure, and cross-linking affect degradability? A review of cell wall model studies.,45(3): 820–831.
Han S H, Yoo S C, Lee B D, An G, Paek N C. 2015. Rice FLAVIN-BINDING, KELCH REPEAT, F-BOX 1 () promotes flowering independent of photoperiod.,,38(12): 2527–2540.
Hansen G, Das A, Chilton MD. 1994. Constitutive expression of the virulence genes improves the efficiency of plant transformation by.,91(16): 7603–7607.
Hansen G, Shillito R D, Chilton MD. 1997. T-strand integration in maize protoplasts after codelivery of a T-DNA substrate and virulence genes., 94: 11726–11730.
Hellwig S, Drossard J, Twyman R M, Fischer R. 2004. Plant cell cultures for the production of recombinant proteins.,22(11): 1415–1422.
Hempel F, Bozarth A S, Lindenkamp N, Klingl A, Zauner S, Linne U, Steinbüchel A, Maier U G. 2011. Microalgae as bioreactors for bioplastic production.,10(1): 81.
Hiei Y, Ohta S, Komari T, Kumashiro T. 1994. Efficient transformation of rice (L.) mediated byand sequence analysis of the boundaries of the T-DNA.,6(2): 271–282.
Hiei Y, Komari T, Kubo T. 1997. Transformation of rice mediated by.,35: 205–218.
Hiei Y, Ishida Y, Komari T. 2014. Progress of cereal transformation technology mediated by.,5: 628.
Hoffmann MC, Pfänder Y, Tintel M, Masepohl B. 2017. Bacterial PerO permeases transport sulfate and related oxyanions.,199(14): 83–117.
Horsch R B, Fraley R T, Rogers S G, Sanders P R, Lloyd A, Hoffmann N. 1984. Inheritance of functional foreign genes in plants.,223: 496–498.
Hou C X, Lv T, Zhan Y H, Peng Y Y, Huang Y Y, Jiang D A, Weng X Y. 2015. Overexpression of the RIXI xylanase inhibitor improves disease resistance to the fungal pathogen,, in rice.,,120(1): 167–177.
Isikgor F H, Becer C R. 2015. Lignocellulosic biomass: A sustainable platform for the production of bio-based chemicals and polymers.,6: 4497–4559.
Ithape D M, Maharana M, Tripathy Swapan K. 2017. Scope of genetic transformation in Sugarcane: A review.,8(1): 1–7.
Izawa T, Shimamoto K. 1996. Becoming a model plant: The importance of rice to plant science.,1(3): 95–99.
Jiang H M, Doerge R W, Gelvin S B. 2003. Transfer of T-DNA and vir proteins to plant cells byinduces expression of host genes involved in mediating transformation and suppresses host defense gene expression.,35(2): 219–236.
Joersbo M, Okkels F T. 1996. A novel principle for selection of transgenic plant cells: Positive selection.,16: 219–221.
Kang J M, Turano F J. 2003. The putative glutamate receptor 1.1 (AtGLR1.1) functions as a regulator of carbon and nitrogen metabolism in.,100(11): 6872–6877.
Karthikeyan A, Pandian S T K, Ramesh M. 2009. High frequency plant regeneration from embryogenic callus of a popularrice (L.)., 15(4): 371–375.
Koetle M J, Finnie J F, Balázs E,van Staden J. 2015. A review on factors affecting the-mediated genetic transformation in ornamental monocotyledonous geophytes.,98: 37–44.
Koetle M J, Baskaran P, Finnie J F, Soos V, Balázs E,van Staden J. 2017. Optimization of transientexpression of-mediated transformation inHilliard using sonication and.,111: 307–312.
Komiya R, Ikegami A, Tamaki S, Yokoi S, Shimamoto K. 2008.andare essential for flowering in rice.,135(4): 767–774.
Komiya R, Yokoi S, Shimamoto K. 2009. A gene network for long-day flowering activatesencoding a mobile flowering signal in rice.,136(20): 3443–3450.
Koncz C, Schell J. 1986. The promoter of TL-DNA genecontrols the tissue-specific expression of chimaeric genes carried by a novel type ofbinary vector.,204(3): 383–396.
Koziel M G, Beland G L, Bowman C, Carozzi N B, Crenshaw R, Crossland L, Dawson J, Desai N, Hill M, Kadwell S, Launis K, Lewis K, Maddox D, McPherson K, Meghji M R, Merlin E, Rhodes R, Warren G W, Wright M, Evola S V. 1993. Field performance of elite transgenic maize plants expressing an insecticidal protein derived from.,11(2): 194–200.
Krishnan S R, Priya A M, Ramesh M. 2013. Rapid regeneration and ploidy stability of ‘cv IR36’rice (L.) confers efficient protocol forcallus organogenesis andmediated transformation.,54: 47.
Kumria R, Waie B, Rajam M.2001. Plant regeneration from transformed embryogenic callus of an eliterice via.,,67(1): 63–71.
Lacroix B, Zaltsman A, Citovsky V. 2011. Host factors involved in genetic transformation of plant cells by.: Neal Stewart Jr C, Touraev A, Tzfira T. Plant Transformation Technologies. John Wiley & Sons: 1–29.
Laurenceau R, Péhau-Arnaudet G, Baconnais S, Gault J, Malosse C, Dujeancourt A, Campo N, Chamot-Rooke J, Le Cam E, Claverys JP, Fronzes R. 2013. A type IV pilus mediates DNA binding during natural transformation in.,9(6): e1003473.
Lee YS, An G. 2015.controls flowering time by modulating rhythmic flowering time regulators preferentially under short day in rice.,58(2): 137–145.
Li J, Zhu S H, Song X W, Shen Y, Chen H M, Yu J, Yi K K, LiuY F, Karplus V J, Wu P, Deng X W. 2006. A rice glutamate receptor-like gene is critical for the division and survival of individual cells in the root apical meristem.,18(2): 340–349.
Liang Z, Chen K L, Li T D, Zhang Y, Wang Y P, Zhao Q, Liu J X, Zhang H W, Liu C M, Ran Y D, Gao C X. 2017. Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes.,8: 1–5.
Lippincott J A, Lippincott B B. 1978. Cell walls of crown-gall tumors and embryonic plant tissues lackadherence sites.,199: 1075–1078.
Liu J P, Zhang C C, Wei C C, Liu X, Wang M G, Yu F F, Xie Q, Tu J M. 2016. The RING finger ubiquitin E3 ligase OsHTAS enhances heat tolerance by promoting H2O2-induced stomatal closure in rice.,170(1): 429–443.
Lorenz M G, Wackernagel W. 1994. Bacterial gene transfer by natural genetic transformation in the environment.,58(3): 563–602.
Lörz H, Baker B, Schell J. 1985. Gene transfer to cereal cells mediated by protoplast transformation.,199(2): 178–182.
Lowe K, Wu E, Wang N, Hoerster G, Hastings C, Cho MJ, Scelonge C, Lenderts B, Chamberlin M, Cushatt J, Wang L J, Ryan L, Khan T, Chow-Yiu J, Hua W, Yu M, Banh J, Bao Z M, Brink K, Igo E, Rudrappa B, Shamseer P M, Bruce W, Newman L, Shen B, Zheng P Z, Bidney D, Carl Falco S, Register J C, Zhao Z Y, Xu D P, Jones T, Gordon-Kamm W. 2016. Morphogenic regulators baby boom and Wuschel improve monocot transformation.,28(9): 1998–2015.
Lucca P, Hurrell R, Potrykus I. 2001. Genetic engineering approaches to improve the bioavailability and the level of iron in rice grains.,102(2): 392–397.
Ma J K C, Drossard J, Lewis D, Altmann F, Boyle J, Christou P, Cole T, Dale P, van Dolleweerd C J, Isitt V, Katinger D, Lobedan M, Mertens H, Paul M J, Rademacher T, Sack M, Hundleby P A C, Stiegler G, Stoger E, Twyman R M, Vcelar B, Fischer R. 2015. Regulatory approval and a first-in-human phase I clinical trial of a monoclonal antibody produced in transgenic tobacco plants.,13(8): 1106–1120.
Mafakheri H, Taghavi S M, Banihashemi Z, Osdaghi E,Lamichhane J R. 2017. Pathogenicity, host range and phylogenetic position ofspecies associated with sugar beet crown gall outbreaks in Southern Iran.,147(3): 721–730.
Manimaran P, Kumar G R, Reddy M R, Jain S, Rao T B, Mangrauthia S K, Sundaram R M, Ravichandran S, Balachandran S M. 2013. Infection of early and young callus tissues ofrice BPT5204 enhances regeneration and transformation efficiency.,20(6): 415–426.
Mann D G J, LaFayette P R, Abercrombie L L, King Z R, Mazarei M, Halter M C, Poovaiah C R, Baxter H, Shen H, Dixon R A, Parrott W A, Neal Stewart Jr C. 2012. Gateway-compatible vectors for high-throughput gene functional analysis in switchgrass (L.) and other monocot species.,10(2): 226–236.
Martins P K, Ribeiro AP, da Cunha B A D B, Kobayashi A K, Molinari H B C. 2015. A simple and highly efficient-mediated transformation protocol for.,6: 41–44.
Matzke M A, Matzke A J. 1995. How and why do plants inactivate homologous (trans) genes?,107(3): 679–685.
Mohanty A, Sarma N P, Tyagi A K. 1999.-mediated high frequency transformation of an eliterice variety Pusa Basmati 1 and transmission of the transgenes to R2progeny.,147(2): 127–137.
Mulwa R M S, Norton M A, Farrand S K, Skirvin R M. 2015.-mediated transformation and regeneration of transgenicChancellorwine grape plants expressing thegene.,46(3): 110–115.
Opabode J T. 2006.-mediated transformation of plants: Emerging factors that influence efficiency.,1(1): 12–20.
Ou-Lee TM, Turgeon R, Wu R. 1986. Expression of a foreign gene linked to either a plant-virus or a Drosophila promoter, after electroporation of protoplasts of rice, wheat, and sorghum.,83(18): 6815–6819.
Ozawa K, Wakasa Y, Ogo Y, Matsuo K, Kawahigashi H, Takaiwa F. 2012. Development of an efficient-mediated gene targeting system for rice and analysis of rice knockouts lacking granule-bound starch synthase (Waxy) and β1, 2-xylosyltransferase.,53(4): 755–761.
Park S Y, Vaghchhipawala Z, Vasudevan B, Lee LY, Shen Y, Singer K, Waterworth W M, Zhang Z J, West C E, Mysore K S, Gelvin S B. 2015.T-DNA integration into the plant genome can occur without the activity of key non-homologous end-joining proteins.,81(6): 934–946.
Pathi K M, Tula S, Huda K M K, SrivastavaV K, Tuteja N. 2013. An efficient and rapid regeneration via multiple shoot induction from mature seed derived embryogenic and organogenic callus of Indian maize (L.).,8(10): e25891.
Prasad K, Kushalappa K, Vijayraghavan U. 2003. Mechanism underlying regulated expression of RFL, a conserved transcription factor, in the developing rice inflorescence.,120(4): 491–502.
Purwestri Y A, Sari R D K, Anggraeni L N, Sasongko A B. 2015.mediated transformation of rolC:: Hd3a-GFP in black rice (L. cv. Cempo Ireng) to promote early flowering.,14: 469–473.
Rahman Z A, Seman Z A, Basirun N, Julkifle A L, Zainal Z,Subramaniam S. 2011. Preliminary investigations of-mediated transformation inrice MR219 embryogenic callus usinggene.,10: 7805–7813.
Rao K V, Rathore K S, Hodges T K, Fu X, Stoger E, Sudhakar D, Williams S, Christou P, Bharathi M, Bown D P, Powell K S, Spence J, Gatehouse A M R, Gatehouse J A. 1998. Expression of snowdrop lectin (GNA) in transgenic rice plants confers resistance to rice brown planthopper.,15(4): 469–477.
Rashid B, Tariq M, Khalid A, Shams F, Ali Q, AshrafF, Ghaffar I, Khan M I, Rehman R, Husnain T. 2017. Crop improvement: New approaches and modern techniques.,8(3): 18–30.
Rathus C, Bower R, Birch R G. 1993. Effects of promoter, intron and enhancer elements on transient gene expression in sugar-cane and carrot protoplasts.,23(3): 613–618.
Ravikumar G, Manimaran P, Voleti S R, Subrahmanyam D, Sundaram R M, Bansal K C, Viraktamath B C, Balachandran S M. 2014. Stress-inducible expression oftranscription factor greatly improves drought stress tolerance in transgenicrice.,23(3): 421–439.
Reddy S S S, Singh B, Peter A J, Rao T V. 2018. Production of transgenic local rice cultivars (L.) for improved drought tolerance usingmediated transformation.,25(8): 1535–1545.
Reddy S S S, Singh B, Peter A J, Rao T V. 2019. Genetic transformation ofrice varieties involvinggene for improved abiotic stress tolerance., 26(2): 294–300.
Sack M, Hofbauer A, Fischer R, Stoger E. 2015. The increasing value of plant-made proteins.,32: 163–170.
Saharan V, Yadav R C, Yadav N R, Ram K. 2004. Studies on improved-mediated transformation in tworice (L.).,3(11): 572–575.
Sahoo K K, Tripathi A K, Pareek A, Sopory S K, Singla-Pareek S L. 2011. An improved protocol for efficient transformation and regeneration of diverserice cultivars.,7: 49.
Sahoo R K, Tuteja N. 2012. Development of-mediated transformation technology for mature seed-derived callus tissues ofrice cultivar IR64.,3(2): 123–128.
Saidu H, Mohammed S. 2013. Plant response to abiotic stresses: From the perspectives of gene expression.,8(9):81–91.
Saika H, Toki S. 2010. Mature seed-derived callus of the modelrice variety Kasalath is highly competent in-mediated transformation., 29(12): 1351–1364.
Sarangi S, Ghosh J, Bora A, Das S, Mandal A B. 2011.-mediated genetic transformation ofrice varieties involving Am-SOD gene.,10(1): 9–18.
Schoonbeek H J, Wang H H, Stefanato F L, Craze M, BowdenS, Wallington E, Zipfel C, Ridout C J. 2015.EF-Tu receptor enhances bacterial disease resistance in transgenic wheat.,206(2): 606–613.
Shi J, Gao H, Wang H, Lafitte H R, Archibald R L, Yang M, Hakimi S M, Mo H, Habben J E. 2017. ARGOS8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions.,15(2): 207–216.
Shrawat A K, Lörz H. 2006.-mediated transformation of cereals: A promising approach crossing barriers.,4(6): 575–603.
Snell K D, Peoples OP. 2009. PHA bioplastic: A value-added coproduct for biomass biorefineries.,3(4): 456–467.
Sood P, Bhattacharya A, Sood A. 2011. Problems and possibilities of monocot transformation.,55(1): 1–15.
Streatfield S J. 2007. Approaches to achieve high-level heterologous protein production in plants.,5(1): 2–15.
Svitashev S, Young J K, Schwartz C, Gao H R, Falco S C, Cigan A M. 2015. Targeted mutagenesis, precise gene editing and site-specific gene insertion in maize using Cas9 and guide RNA.,169: 931–945.
Tabashnik B E, Gassmann A J, Crowder D W, Carriére Y. 2008. Insect resistance to Bt crops: Evidence versus theory.,26(2): 199–202.
Tan L W, Rahman Z A, Goh HH, Hwang DJ, Ismail I, Zainal Z. 2017. Production of transgenic rice (cv. MR219) overexpressinggene through-mediated transformation.,46(5): 703–711.
Thirumurugan T, Ito Y, Kubo T, Serizawa A, Kurata N. 2008. Identification, characterization and interaction of HAP family genes in rice.,279(3): 279–289.
Tie W W, Zhou F, Wang L, Xie W B, Chen H, Li X H, Lin Y J. 2012. Reasons for lower transformation efficiency inrice using-mediated transformation: Lessons from transformation assays and genome-wide expression profiling.,78: 1–18.
Toriyama K, Arimoto Y, Uchimiya H, Hinata K. 1988. Transgenic rice plants after direct gene transfer into protoplasts.,6(9): 1072–1074.
Tripathi R, Bisht H, Singh R. 2010. Effect of acetosyringone and callus age on transformation for scutellum-derived callus of rice.,1(4): 163–170.
Tzfira T, Li J, Lacroix B, Citovsky V. 2004.T-DNA integration: Molecules and models.,20(8): 375–383.
Tzfira T, Citovsky V. 2006.-mediated genetic transformation of plants: Biology and biotechnology.,17(2): 147–154.
Ullah H, Ullah I, Jadoon S A, Rashid H. 2007. Tissue culture techniques for callus induction in rice.,23(1): 81–86.
Uzé M, Potrykus I, Sautter C. 2000. Factors influencing T-DNA transfer fromto precultured immature wheat embryos (L.).,28(2): 17–23.
Vain P. 2007. Thirty years of plant transformation technology development.,5(2): 221–229.
van der Graaff E, den Dulk-Ras A, Hooykaas P J J. 1996. Deviating T-DNA transfer fromto plants.,31(3): 677–681.
Veluthambi K, Gupta A K, Sharma A. 2003. The current status of plant transformation technologies.,84(3): 368–380.
Vogel J. 2008. Unique aspects of the grass cell wall.,11(3): 301–307.
Wang C G, Hu Z L, Lei A P, Jin B H. 2010. Biosynthesis of poly-3-hydroxybutyrate (PHB) in the transgenic green alga.,46(2): 396–402.
Wendt T, Doohan F, Mullins E.2012. Production of-resistant potato () utilisingOV14.,21(3): 567–578.
Wright M, Dawson J, Dunder E, Suttie J, Reed J, Kramer C, Chang Y, Novitzky R, Wang H, Artim-Moore L. 2001. Efficient biolistic transformation of maize (L.) and wheat (L.) using the phosphomannose isomerase gene,, as the selectable marker.,20(5): 429–436.
Wright T R, Shan G, Walsh T A, Lira J M, Cui C, Song P, Zhuang M, Arnold NL, Lin G, Yau K, Russell S M, Clcchillo R M, Peterson M A, Simpson D M, Zhou N, Ponsamuel J, Zhang Z. 2010. Robust crop resistance to broadleaf and grass herbicides provided by aryloxyalkanoate dioxygenase transgenes.,107: 20240–20245.
Xing SC, Li F, Guo QF, Liu DR, Zhao XX, Wang W. 2009. The involvement of an expansin genefrom wheat in regulating plant cell growth.,53(3): 429–434.
Xu R F, Qin R Y, Li H, Li D D, Li L, Wei P C, Yang J B. 2017. Generation of targeted mutant rice using a CRISPR-Cpf1 system.,15(6): 713–717.
Xue W Y, Xing Y Z, Weng X Y, Zhao Y, Tang W J, Wang L, Zhou H J, Yu S B, Xu C G, Li X H, Zhang Q F. 2008. Natural variation inis an important regulator of heading date and yield potential in rice.,40(6): 761–767.
Yang Y, Peng Q, Chen GX, Li XH, Wu CY. 2013.is involved in circadian clock regulation for promoting flowering under long-day conditions in rice.,6(1): 202–215.
Yao JL, Cohen D, Atkinson R, Richardson K, Morris B. 1995. Regeneration of transgenic plants from the commercial apple cultivar Royal Gala.,14(7): 407–412.
Yaqoob U, Kaul T, Nawchoo I A. 2017. Development of an efficient protocol formediated transformation of some recalcitrantrice varieties.,22(3): 346–353.
Yookongkaew N, Srivatanakul M, Narangajavana J. 2007. Development of genotype-independent regeneration system for transformation of rice (ssp.).,120(2): 237–245.
Yoon D H, Lee S S, Park H J, Lyu J I, Chong W S, Liu J R, Kim BG, Ahn J C, Cho H S. 2015. Overexpression ofincreases tolerance to cold stress and enhances grain yield in rice ().,67(1): 69–82.
Zaidi M A, Narayanan M, Sardana R, Taga I, Postel S, Johns R, McNulty M, Mottiar Y, Mao J, Loit E, Altosaar I. 2006. Optimizing tissue culture media for efficient transformation of differentrice genotypes.,4(2): 563–575.
Zhang H M, Yang H, Rech E L, Golds T J, Davis A S, Mulligan B J, Cocking E C, Davey M R. 1988. Transgenic rice plants produced by electroporation-mediated plasmid uptake into protoplasts.,7(6): 379–384.
Zhang HX, Blumwald E. 2001. Transgenic salt-tolerant tomato plants accumulate salt in foliage but not in fruit.,19(8): 765–768.
Zhao J, Chen H Y, Ren D, Tang H W, Qiu R, Feng J L, Long Y M, Niu B X, Chen D P, Zhong T Y, Liu Y G, Guo J X. 2015. Genetic interactions between diverged alleles of() and()/() control differential heading and contribute to regional adaptation in rice ()., 208(3): 936–948.
Zhao W N, Zheng S S, Ling HQ. 2011. An efficient regeneration system and-mediated transformation of Chinese upland rice cultivar Handao297.,,106: 475.
Zhu YJ, Fan YY, Wang K, Huang DR, Liu WZ, Ying JZ, Zhuang JY. 2017. Rice flowering locusplays an important role in heading date influencing yield traits in rice.,7: 4918.
Ziemienowicz A. 2014.-mediated plant transformation: Factors, applications and recent advances.,3(4): 95–102.
Zuniga-Soto E, Mullins E, Dedicova B. 2015. Ensifer-mediated transformation: An efficient non-protocol for the genetic modification of rice.,4: 600.
Copyright © 2019, China National Rice Research Institute. Hosting by Elsevier B V
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/)
Peer review under responsibility of China National Rice Research Institute
http://dx.doi.org/10.1016/j.rsci.2019.04.001
15 May 2018;
10 September 2018
Zaidah Rahmat (zaidahrahmat@utm.my)
(Managing Editor: Li Guan)
- Rice Science的其它文章
- Molecular Markers and Candidate Genes for Thermo-Sensitive Genic Male Sterile in Rice
- Morpho-Physiological Changes in Roots of Rice Seedling upon Submergence
- Biochemical and Metabolomics on Rice Cultivars
- Classification and Identification of indica P/TGMS Lines in China
- Comparative Efficacies of Next-Generation Insecticides Against Yellow Stem Borer and Their Effects on Natural Enemies in Rice Ecosystem
- Morphological and Molecular Characterization of Novel Salt-tolerant Rice Germplasms from the Philippines and Bangladesh

