Occurrence of Escherichia coli virulence genes in feces of wild birds from Central Italy
Fabrizio Bertelloni, Errica Lunardo, Guido Rocchigiani, Renato Ceccherelli, Valentina Viginia Ebani✉
1Department of Veterinary Science, University of Pisa, viale delle Piagge 2, 56124 Pisa, Italy
2CRUMA-LIPU, via delle Sorgenti 430, 57121 Livorno, Italy
Keywords:Wild birds Enteropathogenic Escherichia coli Enterohemorrhagic Escherichia coli Shiga-toxins producing Escherichia coli
ABSTRACT Objective: To investigate the potential role of wild birds as fecal spreaders of enteropathogenic,enterohemorrhagic and Shiga-toxins producing Escherichia coli (E. coli ), enteropathogenic E.coli, enterohemorrhagic E. coli and Shiga toxin-producing E. coli strains.Methods: Fecal samples collected from 121 wild birds of different orders and species were submitted to molecular analyses. In particular, eaeA encoding intimin, hlyA encoding for hemolysin, stx1 and stx2 genes encoding Shiga-toxins 1 and 2, respectively, were investigated.Results: Overall, 21(17.35%) fecal samples resulted positive for at least one of the investigated genes. In detail, 12(9.91%) samples were positive for eaeA, 10(8.26%) for stx1, 4(3.31%) for hylA and 1(0.83%) for stx2. An owl (Athene noctua) positive for the four investigated genes suggesting that it harbored a STEC strain. However, virulence genes characterizing EPEC, and EHEC strains were mainly found among seagulls, waterfowl and feral pigeons.Conclusions: Seagulls, waterfowl and feral pigeons, which frequently reach and contaminate rural, urban and peri-urban areas with their droppings, may be important sources of E. coli infection for other animals and humans.
1. Introduction
Escherichia coli (E. coli) is an opportunistic Gram negative bacterium presenting in the normal gut microflora of human and animals, even though some strains have been related to a wide range of diseases of veterinary and human concern[1].
Enterohemorrhagic E. coli (EHEC) strains are responsible for gastrointestinal disease in humans including diarrhea, hemorrhagic colitis and hemolytic uremic syndrome (HUS), which is the main cause of acute renal failure in children[2]. One of the most important virulence factors of EHEC is hemolysin, which contributes to the pathogenesis by different mechanisms such as adhesion, induction of pro-inflammatory reactions and epithelial and endothelial cell damage[3].
Some EHEC strains produce Shiga-toxins Stx1 and Stx2, which represent virulence factors causing intestinal cell death and consequent diarrhea[4]. These strains, named Shiga toxin-producing E. coli (STEC) or verocytotoxic E. coli (VTEC), are important zoonotic agents. Among them, O157:H7 is the most common serotype encountered in hemorrhagic colitis and HUS cases, even though other non-O157 E. coli serogroups have been involved in human infections[5].
All EHEC express intimin, constitutes an additional virulence factor mediate the intimate attachment of bacteria to epithelial cells and stimulate mucosal immune responses and intestinal crypt hyperplasia, resulting in the characteristic attaching and effacing (A/E) lesions on intestinal epithelial cells[1,2].
E. coli strains producing intimin, but not hemolysin, are termed Enteropathogenic E. coli (EPEC) and are related to potentially fatal diarrhea, affecting mainly children in developing countries. Bacterial adherence to enterocytes mediated by intimin induce microvilli loss,which could eventually lead to diarrhea[6].
Domestic ruminants, mainly cattle, are considered the main reservoirs of STEC, even though these strains have been found among several domestic and wild mammals’ species, too[7].
Following the first report of STEC infection in wild birds in 1997[8], some studies have been carried out to verify the potential role of birds, including poultry, as spreaders of pathogenic E. coli worldwide[6,9-17]. Detection of these pathogens in avian population suggests that birds, together with mammals, may be direct or indirect source of infection for humans.
E. coli O157:H7 was isolated from layer hens and from pigeons in Italy[18,19]. However, to the best of our knowledge, data about EPEC,EHEC and STEC infections in wild avian population in Italy are not available.
The aim of the present study was to investigate the potential role of wild birds as fecal spreaders of EPEC, EHEC and STEC strains by searching the genes coding for the virulence factors intimin,hemolysin and Shiga-toxins in the feces of free-living birds of several species and from different environments.
2. Materials and methods
2.1. Animal samples
From January to December 2016, gut samples were collected from 121 free-roaming wild birds. In particular, 56 samples were collected from hunted animals. These birds have been hunted during the hunting season in different wet areas of Central Italy. While the evisceration was performed by the hunters, intestine was collected and sent to the laboratory of the Avian Pathology Section (APS) of the Department of Veterinary Science (University of Pisa), at 4 ℃.Furthermore, 65 samples were collected from birds dead at a bird recovery center located in Central Italy. During the necropsies,a portion of terminal intestine, approximatively from caeca to cloaca, were collected from each bird and stored at 4 ℃ until the investigations.
2.2. Ethic statement
Regularly hunted and naturally dead birds were used in the study.No birds were sacrificed for the study.
2.3. Molecular analyses
DNA was extracted from about 25 mg of each fecal sample with the commercial kit Tissue Genomic DNA Extraction Kit (Fisher Molecular Biology, Trevose, PA, USA), following the manufacturers’guidelines; once obtained, DNA samples were kept at 4 ℃ until used as template in PCR assays.
Single PCR protocols were performed. No multiplex PCR were executed to allow a better interpretation of the results. Each PCR assay employed a primer pair: stx1F (5’-ATAAATCGCCATTCGTTGACTAC-3’) and stx1R (5’-GAACGCCCACTGAGATCATC-3’) to amplify a 180 bp fragment of stx1 gene encoding Shiga toxins 1, stx2F (5’-GGCACTGTCTGAAACTGCTCC-3’) and stx2R (5’-TCGCCAGTTATCTGACATTCTG-3’) to amplify a 255 bp fragment of stx2 gene encoding Shiga toxins 2, eaeAF(5’-GACCCGGCACAAGCATAAGC-3’) and eaeAR (5’-CCACCTGCAGCAACAAGAGG-3’) to amplify a 384 bp fragment of eaeA gene encoding intimin, and hlyAF (5’-GCATCATCAAGCGTACGTTCC-3’) and hlyAR (5’-AATGAGCCAAGCTGGTTAAGCT-3’) to amplify a 534 bp fragment of hlyA gene encoding for hemolysin. All primers were previously described by Paton and Paton[20].
Each PCR assay was performed in a volume of 50 µL consisting of 25 µL of EconoTaq PLUS 2x Master Mix (Lucigen Corporation,Middleton, Wiskonsin, USA), 0.5 µM of each primer, 3 µL of DNA and distilled water to reach the final volume. The amplifications were carried out in the automated thermal cycler Gene-Amp PCR System 2700 (Perkin Elmer, Norwalk, Connecticut, USA) for 35 cycles. Each cycle consisted of denaturation at 95 ℃ for 1 min,annealing at 60 ℃ for 2 min, extension at 72 ℃ for 1 min; an initial denaturation of 10 min at 95 ℃ and a final extension of 10 min at 72 ℃ were done. PCR assays included a reagent blank as negative control. Quantitative Genomic DNA from O157:H7 E. coli (ATCC®43895DQ TM) was included as positive control to assess the protocols, whereas no positive controls were used in the following amplifications to avoid cross-contaminations. PCR products were analyzed by electrophoresis on 1.5% agarose gel at 100 V for 45 min. PCR Sizer 100 bp DNA Ladder (Norgen Biotek, Thorold,Canada) was used as DNA marker.
3. Results
The species of 56 hunted and 65 dead birds are shown in Figure 1.All hunted birds were in good nutrition conditions and no lesions were observed during the carcasses’ manipulation. The remaining birds were sent to the APS for necropsies, showing fatal traumatic lesions as putative cause of death. No lesions ascribable to infectious and/or parasitic diseases were observed.PCR protocols allowed the amplification and the detection of the four investigated genes fragments when the DNA positive control was tested.

Figure 1. Number of examined birds in relation to the avian species.
Overall, 21(17.35%) fecal samples resulted positive for at least one of the investigated genes. In detail, 12(9.92%) samples were positive for eaeA, 10(8.26%) for stx1, 4(3.31%) for hylA and 1(0.83%) for stx2.
In particular, 8(6.61%) animals were positive only for stx1,8(6.61%) only for eaeA, 1(0.83%) only for hlyA, 2(1.65%) for eaeA and hlyA, 1(0.83%) for stx1 and eaeA, 1(0.83%) for stx1, stx2, eaeA and hlyA.
Considering the examined avian species, positive samples were detected in 8 seagulls [Larus michahellis (L. michahellis)], 5 pigeons[Columba livia (C. livia)], 3 mallard ducks [Anas platyrhynchos (A.platyrhynchos)], 1 common teal [Anas crecca (A. crecca)], 1 grey heron [Ardea cinerea (A. cinerea)], 1 little owl [Athene noctua (A.noctua)], 1 Eurasian wigeon [Anas penelope (A. penelope)] and 1 pintail [Anas acuta (A. acuta)]. Table 1 reports the distribution of positive animals in relation to the detected genetic profiles. Overall,17.35% (21/121) birds were positive for the detected genes, of which 4.13% (5/121) were pigeons, 5.79% (7/121) were waterfowl, 6.61%(8/121) were seagulls, and 0.82% (1/121) were raptors.
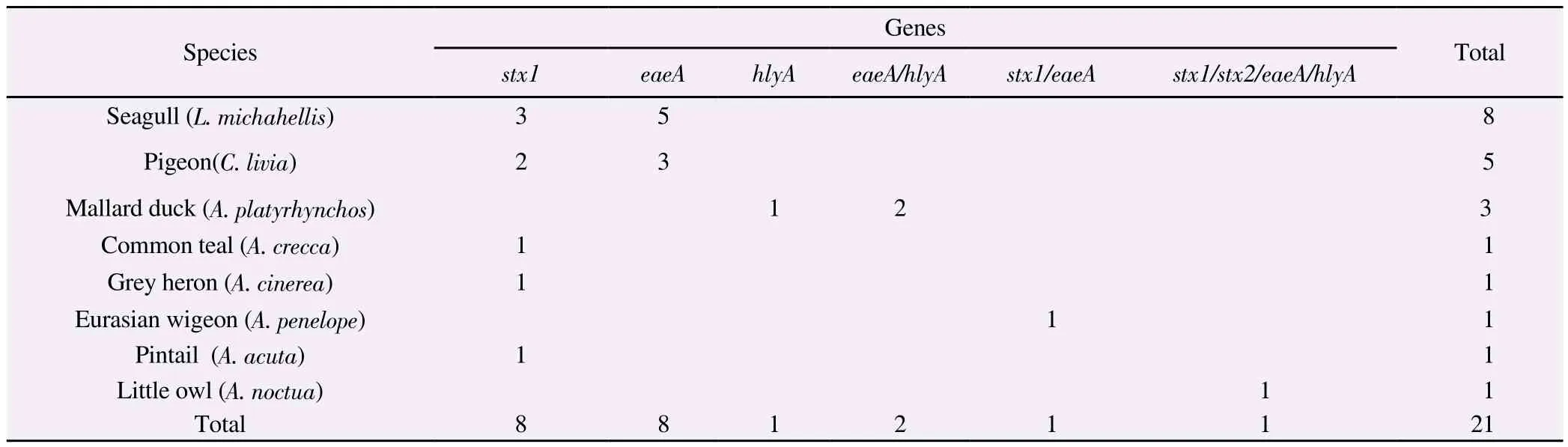
Table 1. Results of PCR assays for the detection of E. coli virulence genes in relation to the animal species.
4. Discussion
The present study was carried out on several bird species found dead or hunted that lived in different environments. Most of them were animals roaming within an aquatic environment, such as seagulls and waterfowl, whereas others usually live in hinterland such as raptors. Moreover, feral pigeons which reach urban, periurban and rural areas were also examined.
The results show that these animals may harbor diarrhea-inducing E. coli strains carrying genes for virulence factors able to determine disease both in animals and humans.
The present investigation found the 8.26% of birds positive for stx1 gene, alone or in combination with other virulence genes. This result is in contrast with a study recently carried out in Brazil that did not detect avian feces positive for stx1 gene[6].
Except in the case of one animal, no positive response for stx2 gene was found. This is a comforting result, considering that stx-2 toxin is more toxic than stx-1 and is often associated with HUS in children[21]. Similar results were obtained by Sanches et al.[6], which found 3/401 (0.75%) stx2-positive birds.
One bird (A. platyrhynchos) was positive only for hlyA gene,whereas two others (A. platyrhynchos) were positive for both hlyA and eaeA genes, suggesting they were spreaders of EHEC strains.The gene eaeA, characterizing EPEC strains, was found alone in eight (6.61%) birds, five seagulls and three pigeons, and in combination with stx1 in one wigeon. This value is in agreement with the study by Sanches et al.[6] that found 5.74% of eaeA-positive wild birds. Prevalence obtained in other investigations is difficult to compare to our results, since the populations examined are different from our study.
However, other authors mainly found eaeA-positive birds among Columbiformes[6,22]. In this study stx, eaeA and hlyA genes were mainly detected in the fecal samples collected from seagulls,waterfowl and pigeons.
Seagulls (L. michahellis) are omnivorous aquatic birds, which usually live along the marine coasts. However, they can be found around lakes and in urban/peri-urban areas. They eat mainly fish,rats, animal carcasses, but more often they feed on human food waste found in garbage bins or in land fills[23].
Waterfowl, including ducks and geese, are largely found in wetlands; they indeed can be found in areas with rivers and lakes, but also small ponds, including those presenting in public or private parks. These animals are able to travel large distances per day dispersing pathogens over wide areas. They are largely involved in the epidemiology of some infectious diseases, such as Avian Influenza and Newcastle Disease, because they may excrete/contract the pathogens in the wetland environments shared by numerous birds[24]. At the same way, geese and ducks can contaminate surface water and pasture with other pathogens,including E. coli. Moreover, the contaminated surface water, if used for irrigation, represents a further source of infection for people.
Waterfowl are the most common hunted birds. If infected by E.coli, they represent a dangerous source of infection for hunters and other people during the carcasses’ manipulation, a procedure often carried out in kitchens.
Positive responses detected among feral pigeons (C. livia) are relevant for the public health, as well. They are synanthropic birds largely present in urban and peri-urban areas and can contaminate,with their droppings, environments where people are also living[25].Moreover, pigeons may be source of infection for other animals,such as birds, mammals and reptiles also in rural areas. In particular, pigeons, such as other wild birds, frequently live around farms and are carriers of pathogens for livestock.
During this investigation, a fecal sample collected from an owl (A. noctua) scored positive for all the investigated genes.Bacteriological examinations were carried out on this sample and an E. coli strain was isolated and typed by cultural and staining characteristics and API 20E System (BioMérieux, Marcy l’Etoile,France). The strain, submitted to molecular analyses, resulted to have the four investigated virulence genes, suggesting that the animal harbored a STEC strain. On the basis of the available literature, this is the first report of STEC infection in an owl, thus no information about possible pathology due to this type of E. coli in owls is known.
Owl is a carnivorous bird of prey that lives mainly on a diet of insects, earthworms, small vertebrates including amphibians,reptiles, birds and small mammals, such as mice, rats, shrews,lagomorphs, voles and moles. This STEC-positive animal could have ingested the pathogen feeding on a prey. This result suggests that STEC strains are circulating, even if not largely, among wildlife, as also demonstrated by other studies that found STEC in Columbiformes[6,22].
In particular, the source of infection for the positive owl could be rodents, which are largely found in rural areas, such as livestock farms. A previous study detected the same STEC strains in feces from rodents and cattle[9].
The obtained results do not clarify whether wild birds can act as spillover hosts or reservoirs for infection. E. coli present in the feces of the investigated birds could be transient or resident bacteria in the intestinal tract. Regardless STEC, EPEC and EHEC strains replicate or not in the avian intestine, the presence of these pathogens in the droppings of birds is a severe threat for health of other animals and humans.
5. Conclusion
This preliminary investigation, even though carried out on a limited number of samples, shows that wild birds can harbor and excrete in their feces pathogenic E. coli strains able to infect and determine disease in other animal species and humans. The highest rates of infection were found among seagulls, waterfowl and feral pigeons, constituting a potential source of E. coli for people,companion animals and livestock, as they live in urban, peri-urban and rural environments. Considering that bacterial prophylactic measures on wild birds are not achievable, environmental hygienic actions are basic to reduce the pathogens’ spreading. Attention should be paid to the waste management, bins and dumps, which could attract pigeons and gulls near to the human environment.Moreover, surface water should not be used for irrigation and watering of the animals and measures against wild birds are necessary to reduce the environment contaminations with avian droppings.
Conflict of interest statement
The authors declare no conflict of interest.
 Asian Pacific Journal of Tropical Medicine2019年3期
Asian Pacific Journal of Tropical Medicine2019年3期
- Asian Pacific Journal of Tropical Medicine的其它文章
- SARS and its treatment strategies
- Phyllanthus acidus (L.) Skeels and Rhinacanthus nasutus (L.) Kurz leaf extracts suppress melanogenesis in normal human epidermal melanocytes and reconstitutive skin culture
- Antihydatic and immunomodulatory effects of Algerian propolis ethanolic extract: In vitro and in vivo study
- Chemical composition of Mentha suaveolens and Pinus halepensis essential oils and their antibacterial and antioxidant activities
- Isolation and structural elucidation of antifungal compounds from Curcuma amada
- Status of intestinal parasitic infections among rural and urban populations, southwestern Iran
